Search Count: 22
 |
Crystal Structure Of Mbp-Fused Bil1/Bzr1 (21-90) In Complex With Double-Stranded Dna Contaning Atcacgtgat
Organism: Serratia sp. (strain fs14), Arabidopsis thaliana, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2022-12-07 Classification: TRANSCRIPTION Ligands: EDO |
 |
Crystal Structure Of Mbp-Fused Bil1/Bzr1 (21-90) In Complex With Double-Stranded Dna Contaning Cacacgtgtg
Organism: Serratia sp. (strain fs14), Arabidopsis thaliana, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2022-12-07 Classification: TRANSCRIPTION Ligands: EDO |
 |
Crystal Structure Of Mbp-Fused Bil1/Bzr1 (21-90) In Complex With Double-Stranded Dna Contaning Tccacgtgga
Organism: Serratia sp. (strain fs14), Arabidopsis thaliana, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-12-07 Classification: TRANSCRIPTION Ligands: EDO |
 |
Crystal Structure Of Mbp-Fused Bil1/Bzr1 (21-90) In Complex With Double-Stranded Dna Contaning Ttcacgtgaa
Organism: Serratia sp. (strain fs14), Arabidopsis thaliana, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-12-07 Classification: TRANSCRIPTION Ligands: EDO |
 |
Crystal Structure Of Mbp-Fused Bil1/Bzr1 (21-90) In Complex With Double-Stranded Dna Contaning Cgcacgtgcg
Organism: Serratia sp. (strain fs14), Arabidopsis thaliana, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2022-12-07 Classification: TRANSCRIPTION Ligands: EDO |
 |
Crystal Structure Of Mbp-Fused Bil1/Bzr1 (21-90) In Complex With Double-Stranded Dna Contaning Gacacgtgtc
Organism: Serratia sp. (strain fs14), Arabidopsis thaliana, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2022-12-07 Classification: TRANSCRIPTION Ligands: EDO |
 |
Crystal Structure Of Mbp-Fused Bil1/Bzr1 (21-90) In Complex With Double-Stranded Dna Contaning Gtcacgtgac
Organism: Serratia sp. (strain fs14), Arabidopsis thaliana, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2022-12-07 Classification: TRANSCRIPTION Ligands: EDO |
 |
Crystal Structure Of Gh121 Beta-L-Arabinobiosidase Hypba2 From Bifidobacterium Longum
Organism: Bifidobacterium longum
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-06-03 Classification: HYDROLASE Ligands: PEG, PGE, EDO, PG4, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2020-04-15 Classification: LIGASE Ligands: ZN, D5U, GOL, EPE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2020-04-15 Classification: LIGASE Ligands: ZN, D60, CL, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2018-07-25 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 8LU |
 |
Mouse Cereblon Thalidomide Binding Domain Complexed With Racemic Thalidomide
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-02-07 Classification: METAL BINDING PROTEIN Ligands: EF2, ZN, SO4 |
 |
Mouse Cereblon Thalidomide Binding Domain Complexed With S-Form Thalidomide
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-02-07 Classification: METAL BINDING PROTEIN Ligands: EF2, ZN, SO4 |
 |
Mouse Cereblon Thalidomide Binding Domain Complexed With R-Form Thalidomide
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-02-07 Classification: METAL BINDING PROTEIN Ligands: 6EL, ZN, SO4 |
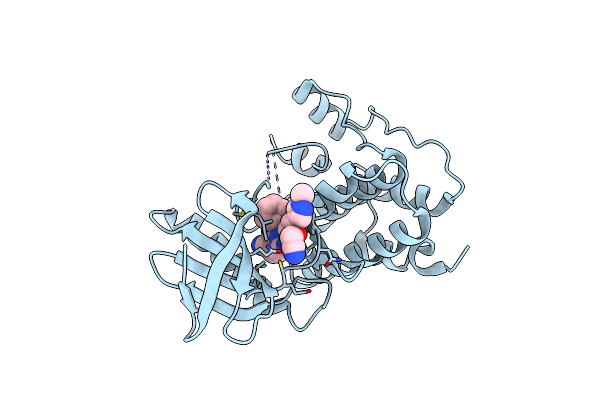 |
Crystal Structure Of Human Mps1 Catalytic Domain In Complex With 6-((3-(Cyanomethoxy)-4-(1-Methyl-1H-Pyrazol-4-Yl)Phenyl)Amino)-2-(Cyclohexylamino)Nicotinonitrile
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-04-08 Classification: TRANSFERASE Ligands: IOD, O38 |
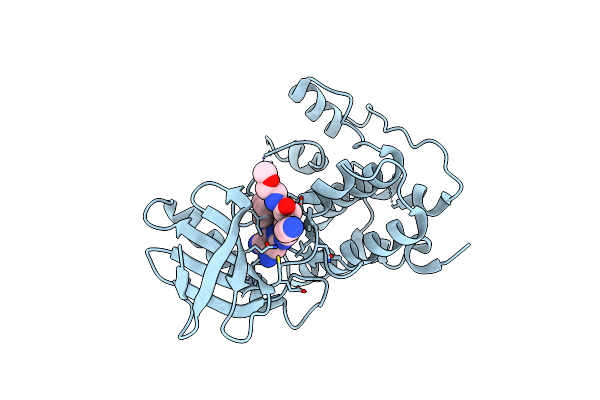 |
Crystal Structure Of Human Mps1 Catalytic Domain In Complex With (E)-3-(4-((6-(((3S,5S,7S)-Adamantan-1-Yl)Amino)-4-Amino-5-Cyanopyridin-2-Yl)Amino)-2-(Cyanomethoxy)Phenyl)-N-(2-Methoxyethyl)Acrylamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2015-04-08 Classification: TRANSFERASE Ligands: O17 |
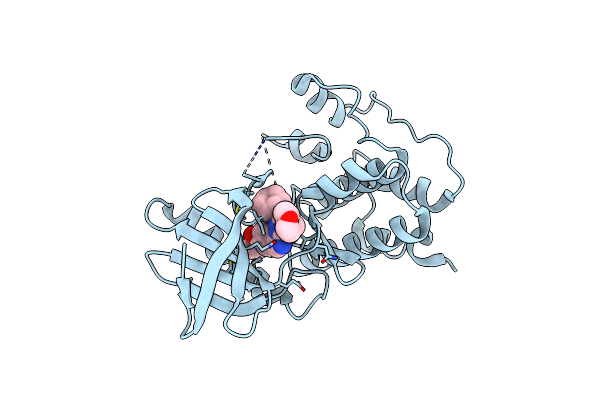 |
Crystal Structure Of Human Mps1 Catalytic Domain In Complex With 4-(6-(Cyclohexylamino)-8-(((Tetrahydro-2H-Pyran-4-Yl)Methyl)Amino)Imidazo[1,2-B]Pyridazin-3-Yl)-N-Cyclopropylbenzamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2015-02-11 Classification: TRANSFERASE Ligands: O43 |
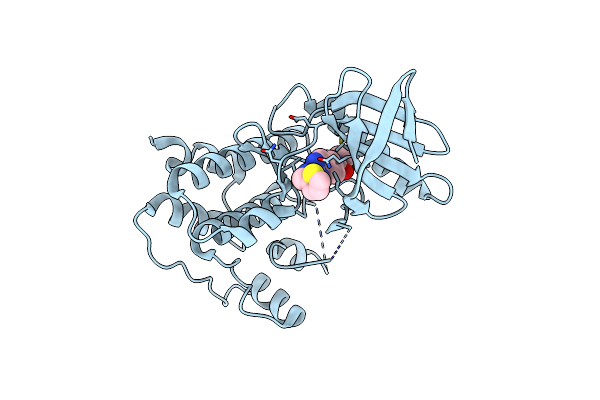 |
Crystal Structure Of Human Mps1 Catalytic Domain In Complex With N-Cyclopropyl-4-(8-((Thiophen-2-Ylmethyl)Amino)Imidazo[1,2-A]Pyrazin-3-Yl)Benzamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-02-11 Classification: TRANSFERASE Ligands: CL, O23 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2013-10-09 Classification: LYASE Ligands: FES, CHD, SAL, EDO |
 |
Crystal Structure Of Human Mps1 Catalytic Domain In Complex With 5-(5-Ethoxy-6-(1-Methyl-1H-Pyrazol-4-Yl)-1H-Indazol-3-Yl)-2-Methylbenzenesulfonamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2013-06-26 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 1O5 |

