Search Count: 193
 |
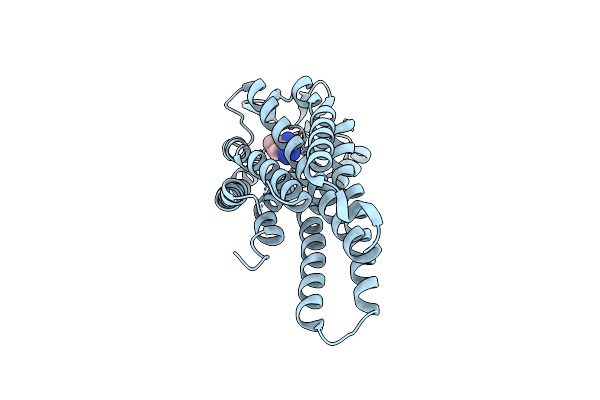
Locally-Refined Structure Of Alpha2A Adrenergic Receptor In Complex With Go Heterotrimer, Scfv16, And Compound Z7149
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: MEMBRANE PROTEIN Ligands: A1CIZ |
 |
Cryoem Structure Of Hca2 Dreadd Gi1 In Complex With Fch-2296413 (Local Refinement)
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: XI9 |
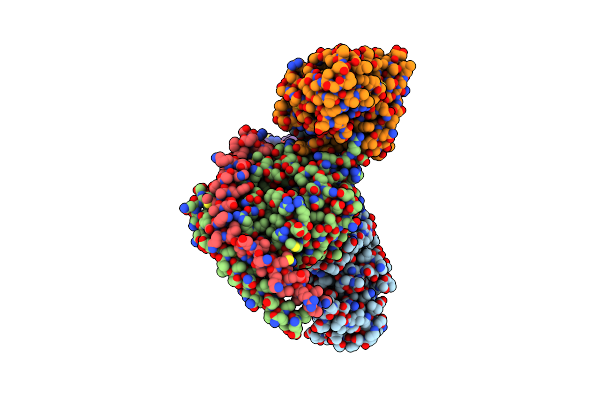 |
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: MEMBRANE PROTEIN Ligands: XOT |
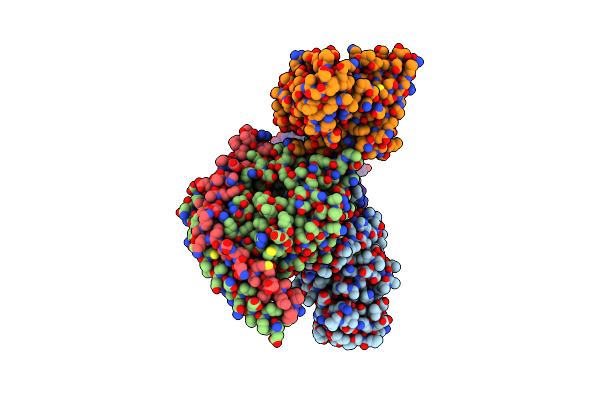 |
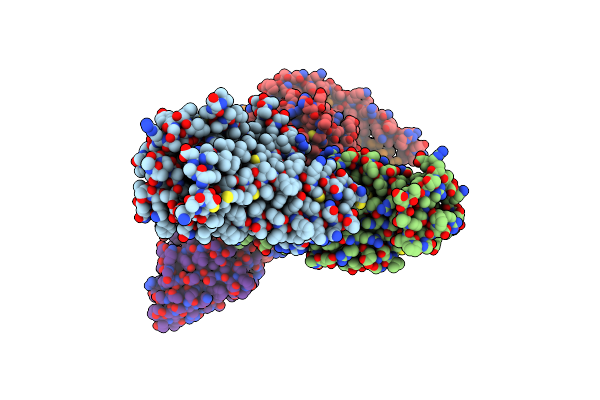
Organism: Homo sapiens, Lama glama, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: MEMBRANE PROTEIN Ligands: XI9 |
 |
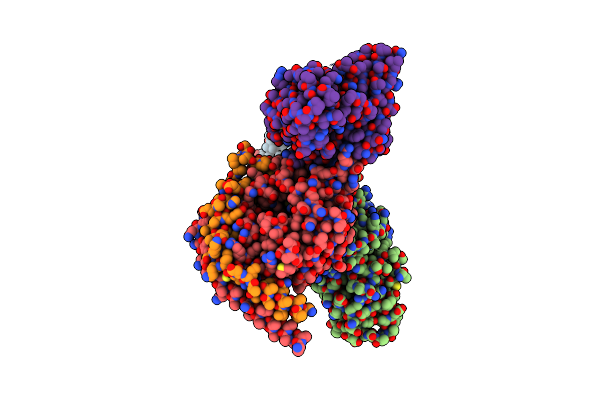
5-Ht2Ar Bound To Lisuride In Complex With A Mini-Gq Protein And An Active-State Stabilizing Single-Chain Variable Fragment (Scfv16) Obtained By Cryo-Electron Microscopy (Cryoem)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-29 Classification: MEMBRANE PROTEIN Ligands: H8G |
 |
5Ht2Ar-Minigq Heterotrimer In Complex With A Novel Agonist Obtained From Large Scale Docking
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-29 Classification: MEMBRANE PROTEIN Ligands: YEQ |
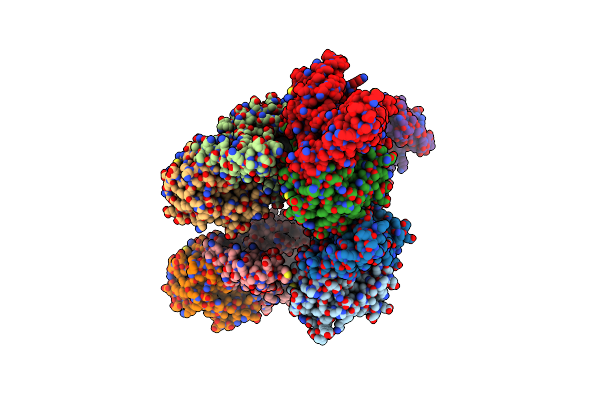 |
Crystal Structure Of Pyrrolysyl-Trna Synthetase From Methanogenic Archaeon Iso4-G1
Organism: Methanogenic archaeon mixed culture iso4-g1
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2023-03-15 Classification: TRANSLATION |
 |
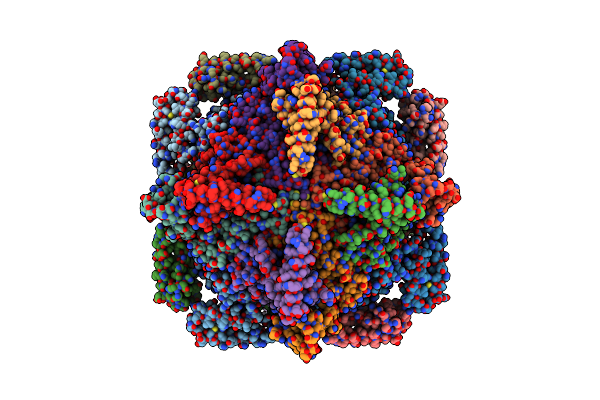
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-06-22 Classification: TRANSFERASE |
 |
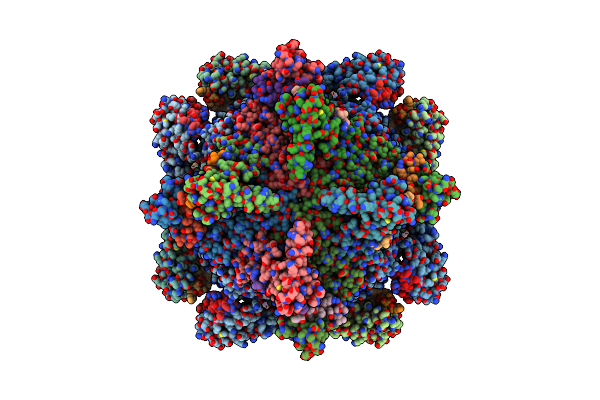
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-03-16 Classification: STRUCTURAL PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-03-16 Classification: STRUCTURAL PROTEIN |
 |
Crystal Structure Of Pyrrolysyl-Trna Synthetase From Methanomethylophilus Alvus
Organism: Candidatus methanomethylophilus alvus
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2019-05-22 Classification: TRANSLATION,LIGASE |
 |
Crystal Structure Of Methanosarcina Mazei Pylrs(Y306A/Y384F) Complexed With Mazzlys
Organism: Methanosarcina mazei jcm 9314
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2019-04-17 Classification: TRANSLATION Ligands: MG, ATP, 9TR, K, PEG, PGE |
 |
Crystal Structure Of Methanosarcina Mazei Pylrs(Y306A/Y384F) Complexed With Mtmdzlys
Organism: Methanosarcina mazei jcm 9314
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2019-04-17 Classification: TRANSLATION Ligands: MG, ATP, 9TU, K |
 |
Crystal Structure Of Methanosarcina Mazei Pylrs(Y306A/Y384F) Complexed With Metzlys
Organism: Methanosarcina mazei jcm 9314
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2019-04-17 Classification: TRANSLATION Ligands: MG, ATP, 9TX, K, PEG |
 |
Crystal Structure Of Methanosarcina Mazei Pylrs(Y306A/Y384F) Complexed With Tco*Lys
Organism: Methanosarcina mazei jcm 9314
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-04-17 Classification: TRANSLATION Ligands: MG, ATP, 9U0, K, PEG |
 |
Crystal Structure Of Methanosarcina Mazei Pylrs(Y306A/Y384F) Complexed With Zaesecys
Organism: Methanosarcina mazei jcm 9314
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-04-17 Classification: TRANSLATION Ligands: MG, ATP, 9U6 |
 |
Crystal Structure Of Methanosarcina Mazei Pylrs(Y306A/Y384F) Complexed With Bcnlys
Organism: Methanosarcina mazei jcm 9314
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-04-17 Classification: TRANSLATION Ligands: MG, ATP, 9U9, K |
 |
Crystal Structure Of Methanosarcina Mazei Pylrs(Y306A/Y384F) Complexed With Pno2Zlys
Organism: Methanosarcina mazei jcm 9314
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2019-04-17 Classification: TRANSLATION Ligands: MG, ATP, 9UC, CIT, K |
 |
Crystal Structure Of Methanosarcina Mazei Pylrs(Y306A/Y384F) Complexed With Pampylys
Organism: Methanosarcina mazei jcm 9314
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2019-04-17 Classification: TRANSLATION Ligands: MG, ATP, 9UF, K, PO4 |
 |
Crystal Structure Of Methanosarcina Mazei Pylrs(Y306A/Y384F) Complexed With Oazzlys
Organism: Methanosarcina mazei jcm 9314
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2019-04-17 Classification: TRANSLATION Ligands: MG, ATP, 9V0, K |

