Search Count: 44
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-02-07 Classification: HYDROLASE Ligands: MG, X5W |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2021-06-23 Classification: IMMUNE SYSTEM Ligands: GD0, NAG |
 |
Crystal Structure Of D-Glucarate Dehydratase From Agrobacterium Tumefaciens Complexed With Magnesium
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-10-30 Classification: ISOMERASE Ligands: CL, MG, NA |
 |
Crystal Structure Of D-Glucarate Dehydratase From Agrobacterium Tumefaciens Complexed With Magnesium, L-Xylarohydroxamate And L-Lyxarohydroxamate
Organism: Agrobacterium fabrum
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2013-09-18 Classification: ISOMERASE Ligands: MG, XYH, LLH, CL, PGE |
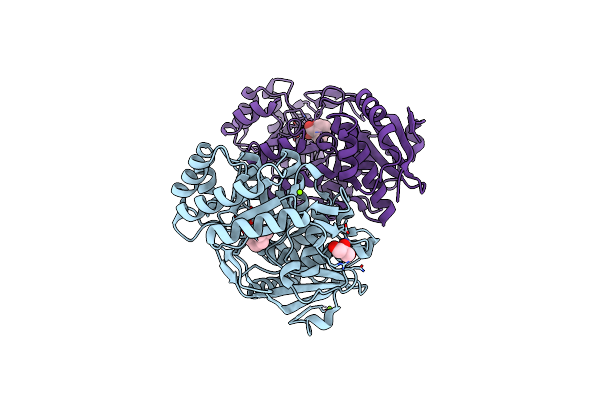 |
Crystal Structure Of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans Pd1222 (Target Nysgrc-012907) With Bound L-Proline Betaine (Substrate)
Organism: Paracoccus denitrificans
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-03-06 Classification: ISOMERASE Ligands: MG, IOD, GOL, PBE |
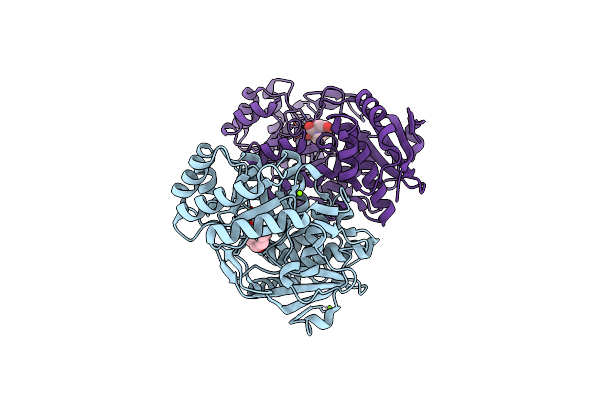 |
Crystal Structure Of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans Pd1222 (Target Nysgrc-012907) With Bound Cis-4Oh-D-Proline Betaine (Product)
Organism: Paracoccus denitrificans
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-02-13 Classification: ISOMERASE Ligands: IOD, MG, 4OP |
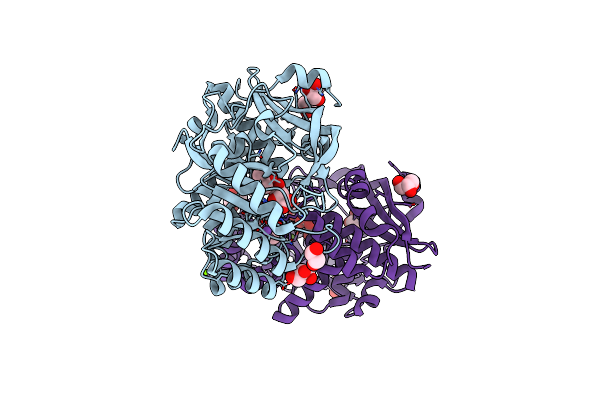 |
Crystal Structure Of D-Glucarate Dehydratase From Agrobacterium Tumefaciens Complexed With Magnesium And L-Tartrate
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-10-24 Classification: ISOMERASE Ligands: TLA, GOL, MG, NA, PGE, EDO |
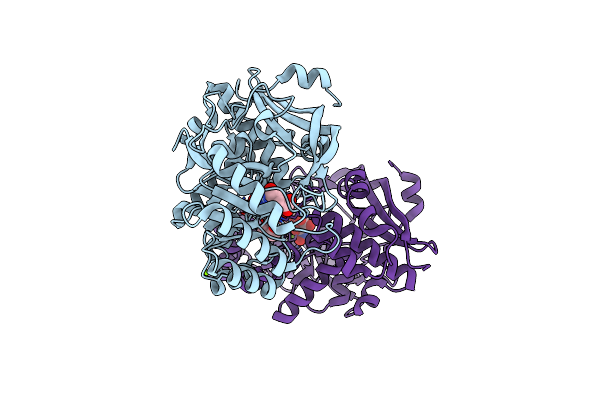 |
Crystal Structure Of D-Glucarate Dehydratase From Agrobacterium Tumefaciens Complexed With Magnesium And L-Lyxarohydroxamate
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-10-24 Classification: ISOMERASE Ligands: MG, LLH, PG4 |
 |
Crystal Structure Of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis Htcc2601, With Bound Mg And L-4-Hydroxyproline Betaine (Betonicine)
Organism: Pelagibaca bermudensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-10-10 Classification: ISOMERASE Ligands: MG, NI, 0XW, IOD, MPD |
 |
Crystal Structure Of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans Pd1222 (Target Nysgrc-012907) With Bound Mg
Organism: Paracoccus denitrificans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-05-02 Classification: ISOMERASE Ligands: MG, UNL |
 |
Crystal Structure Of Dipeptide Epimerase From Methylococcus Capsulatus Complexed With Mg Ion
Organism: Methylococcus capsulatus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2011-05-11 Classification: ISOMERASE Ligands: MG, SO4, GOL |
 |
Crystal Structure Of Dipeptide Epimerase From Methylococcus Capsulatus Complexed With Mg And Dipeptide L-Arg-D-Lys
Organism: Methylococcus capsulatus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2011-04-27 Classification: ISOMERASE Ligands: SO4, ARG, DLY, MG, 1PE |
 |
Crystal Structure Of Nysgrc Enolase Target 200555, A Putative Dipeptide Epimerase From Francisella Philomiragia : Mg Complex
Organism: Francisella philomiragia subsp. philomiragia
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-04-20 Classification: LYASE Ligands: SO4, MG, GOL |
 |
Crystal Structure Of Nysgrc Enolase Target 200555, A Putative Dipeptide Epimerase From Francisella Philomiragia : Mg And Fumarate Complex
Organism: Francisella philomiragia subsp. philomiragia
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-04-20 Classification: ISOMERASE Ligands: SO4, MG, GOL, FUM |
 |
Crystal Structure Of Nysgrc Enolase Target 200555, A Putative Dipeptide Epimerase From Francisella Philomiragia : Complex With L-Ala-L-Glu And L-Ala-D-Glu
Organism: Francisella philomiragia subsp. philomiragia
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-04-20 Classification: ISOMERASE Ligands: ALA, GLU, SO4, GOL, DGL |
 |
Crystal Structure Of Nysgrc Enolase Target 200555, A Putative Dipeptide Epimerase From Francisella Philomiragia : Tartrate And Mg Complex
Organism: Francisella philomiragia subsp. philomiragia
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-04-06 Classification: LYASE Ligands: MG, SO4, TAR, GOL |
 |
Crystal Structure Of Nysgrc Enolase Target 200555, A Putative Dipeptide Epimerase From Francisella Philomiragia : Tartrate Bound, No Mg
Organism: Francisella philomiragia
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-03-30 Classification: METAL BINDING PROTEIN Ligands: SO4, TAR, GOL |
 |
Crystal Structure Of Dipeptide Epimerase From Cytophaga Hutchinsonii Complexed With Mg And Dipeptide D-Ala-L-Val
Organism: Cytophaga hutchinsonii
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2011-02-16 Classification: ISOMERASE Ligands: MG, DAL, VAL |
 |
Crystal Structure Of Dipeptide Epimerase From Cytophaga Hutchinsonii Complexed With Mg And Dipeptide D-Ala-L-Ala
Organism: Cytophaga hutchinsonii
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2011-02-16 Classification: ISOMERASE Ligands: MG, DAL, ALA |
 |
Crystal Structure Of Dipeptide Epimerase From Enterococcus Faecalis V583 Complexed With Mg And Dipeptide L-Arg-L-Tyr
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-10-13 Classification: ISOMERASE Ligands: ARG, TYR, MG |

