Search Count: 13
 |
Crystal Structure Of Sars-Cov-2 Nucleocapsid Phosphoprotein N-Terminal Domain(N-Ntd) In Complex With Ump
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2025-04-23 Classification: VIRAL PROTEIN Ligands: U, PO4 |
 |
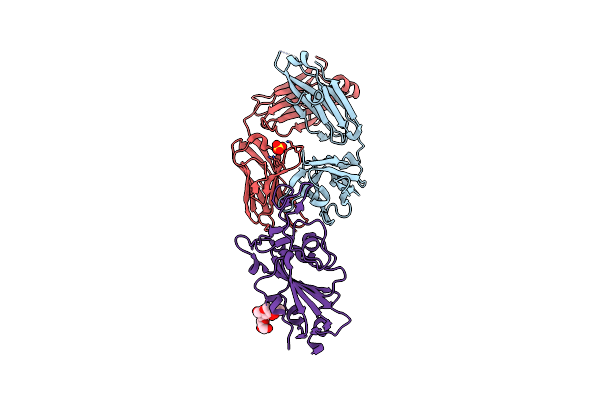
Crystal Structure Of Sars-Cov-2 Spike Receptor-Binding Domain (Rbd) In Complex With Ncv2Sg53 Fab
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2023-04-19 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: SO4, NAG |
 |
Crystal Structure Of Sars-Cov-2 Spike Receptor-Binding Domain (Rbd) In Complex With Ncv2Sg48 Fab
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2023-04-19 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: SO4 |
 |
Crystal Structure Of Sars-Cov-2 Omicron Variant Spike Receptor-Binding Domain (Rbd) In Complex With Ncv2Sg48 Fab
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2023-04-19 Classification: IMMUNE SYSTEM/VIRAL PROTEIN Ligands: SO4 |
 |
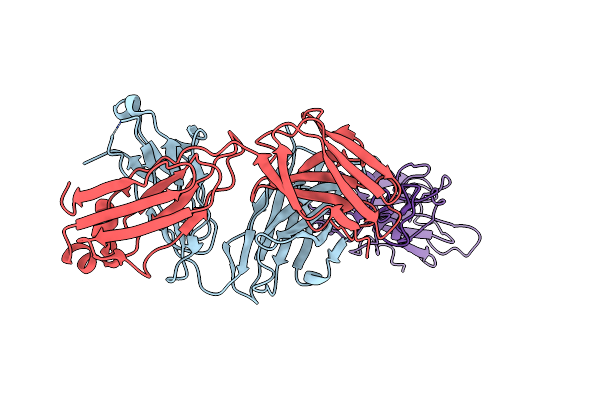
Crystal Structure Of Sars-Cov-2 Delta Variant Spike Receptor-Binding Domain (Rbd) In Complex With Ncv2Sg48 Fab
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2023-04-19 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Crystal Structure Of Sars-Cov-2 Delta Variant Spike Receptor-Binding Domain (Rbd) In Complex With Ncv2Sg53 Fab
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2023-04-19 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
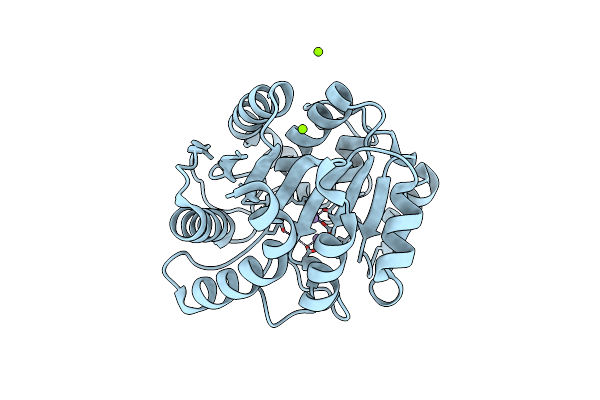
Crystal Structure Of N(Omega)-Hydroxy-L-Arginine Hydrolase In Complex With L-Orn
Organism: Streptomyces lavendulae
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-05-18 Classification: ANTIBIOTIC Ligands: MN, MG, ORN |
 |
Crystal Structure Of C86H-H196S Mutant Of N(Omega)-Hydroxy-L-Arginine Hydrolase
Organism: Streptomyces lavendulae
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2022-05-18 Classification: ANTIBIOTIC Ligands: MN, MG |
 |
Crystal Structure Of N(Omega)-Hydroxy-L-Arginine Hydrolase In Complex With Abh
Organism: Streptomyces lavendulae
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2022-05-18 Classification: ANTIBIOTIC Ligands: MN, MG, ABH |
 |
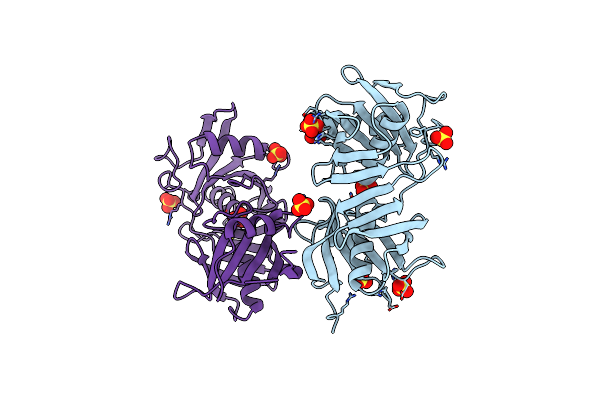
Crystal Structure Of C86H-Y124N-G126H-H196S Mutant Of N(Omega)-Hydroxy-L-Arginine Hydrolase
Organism: Streptomyces lavendulae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-05-18 Classification: ANTIBIOTIC Ligands: MN, MG |
 |
Organism: Streptomyces lavendulae
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2021-12-15 Classification: ISOMERASE Ligands: SO4 |
 |
Organism: Homo sapiens, Murine respirovirus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-08-19 Classification: VIRAL PROTEIN |
 |
Organism: Homo sapiens, Sendai virus (z)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-07-01 Classification: TRANSCRIPTION/VIRAL PROTEIN Ligands: CA |

