Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
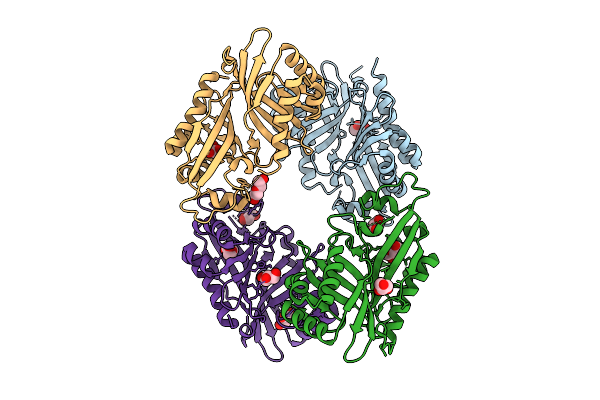 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: GOL, PEG |
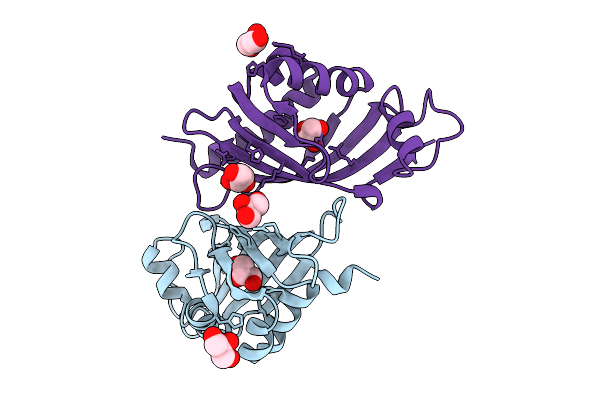 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: GOL |
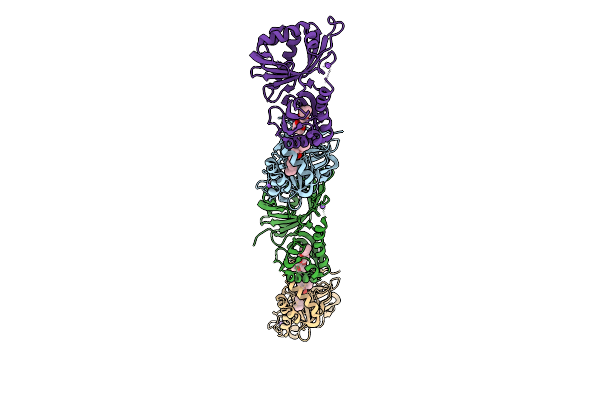 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: A1L6P, NA |
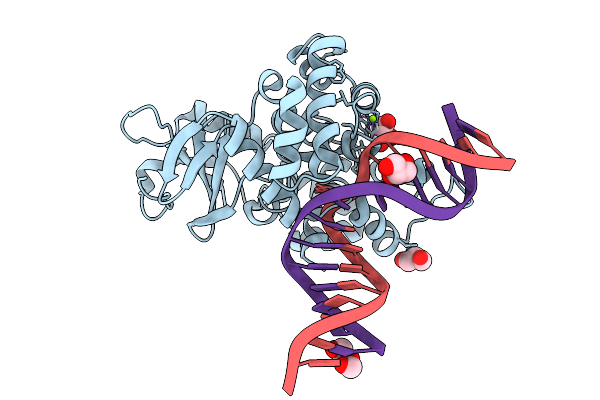 |
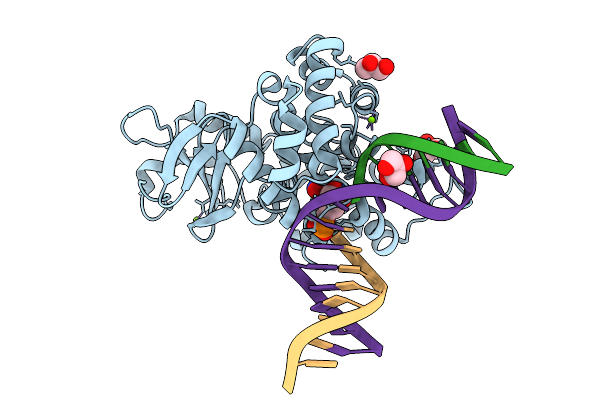
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Substrate 8-Oxoguanine Dna At Ph 8.0 Under 277 K
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: PEG, GOL, MG, NA |
 |
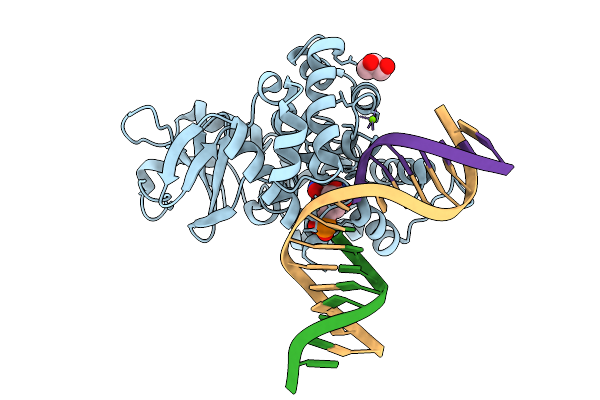
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Reaction Intermediate Derived From The Crystal Soaked Into The Solution At Ph 4.0 Under 277 K For 24 Hourss
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: A1LXK, MG, GOL |
 |
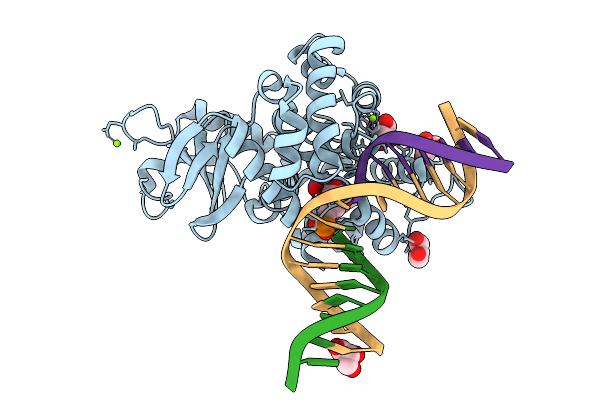
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Reaction Intermediate Derived From The Crystal Soaked Into The Solution At Ph 4.0 Under 277 K For 2.5 Hours
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: A1LXK, MG, GOL |
 |
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Reaction Intermediate Derived From The Crystal Soaked Into The Solution At Ph 4.0 Under 298 K For 3 Weeks
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: A1LXK, GOL, MG, NA |
 |
Solution Structure Of Peptidyl-Prolyl Cis/Trans Isomerase Domain Of Trigger Factor In Complex With Mbp
Organism: Escherichia coli k-12
Method: SOLUTION NMR Release Date: 2018-08-22 Classification: CHAPERONE |
 |
Organism: Escherichia coli (strain k12)
Method: SOLUTION NMR Release Date: 2018-06-13 Classification: CHAPERONE |
 |
Organism: Escherichia coli o157:h7, Escherichia coli (strain k12)
Method: SOLUTION NMR Release Date: 2016-08-24 Classification: CHAPERONE/HYDROLASE |
 |
The Structure Of Chaperone Secb In Complex With Unstructured Phoa Binding Site A
Organism: Escherichia coli (strain 55989 / eaec), Escherichia coli (strain k12)
Method: SOLUTION NMR Release Date: 2016-08-24 Classification: CHAPERONE/HYDROLASE |
 |
The Structure Of Chaperone Secb In Complex With Unstructured Prophoa Binding Site C
Organism: Escherichia coli o157:h7, Escherichia coli (strain k12)
Method: SOLUTION NMR Release Date: 2016-08-24 Classification: CHAPERONE/HYDROLASE |
 |
The Structure Of Chaperone Secb In Complex With Unstructured Prophoa Binding Site D
Organism: Escherichia coli o157:h7, Escherichia coli (strain k12)
Method: SOLUTION NMR Release Date: 2016-08-24 Classification: CHAPERONE/HYDROLASE |
 |
The Structure Of Chaperone Secb In Complex With Unstructured Prophoa Binding Site E
Organism: Escherichia coli o157:h7, Escherichia coli (strain k12)
Method: SOLUTION NMR Release Date: 2016-08-24 Classification: CHAPERONE/HYDROLASE |
 |
The Structure Of Chaperone Secb In Complex With Unstructured Mbp Binding Site D
Organism: Escherichia coli o157:h7
Method: SOLUTION NMR Release Date: 2016-08-24 Classification: CHAPERONE/PROTEIN BINDING |
 |
The Structure Of Chaperone Secb In Complex With Unstructured Mbp Binding Site E
Organism: Escherichia coli o157:h7
Method: SOLUTION NMR Release Date: 2016-08-24 Classification: CHAPERONE/PROTEIN BINDING |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2016-02-17 Classification: ELECTRON TRANSPORT Ligands: HEC |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2016-02-17 Classification: ELECTRON TRANSPORT Ligands: HEC |
 |
Nmr Structure Of E. Coli Trigger Factor In Complex With Unfolded Phoa220-310
|
 |