Search Count: 41
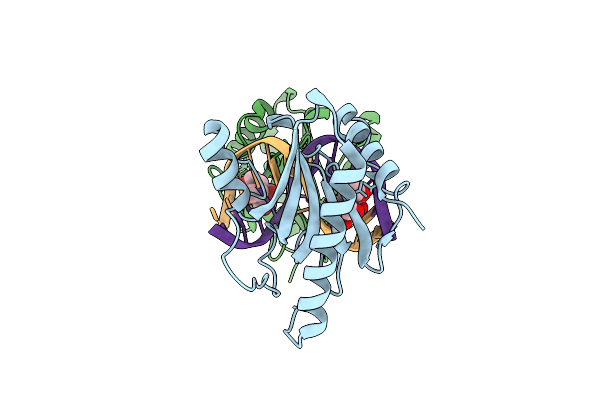 |
Structure Of Dna Binding Domain Of Mcrbc Endonuclease Bound To Dna: Y41F-L68F Double Mutant
Organism: Escherichia coli k-12, Dna molecule
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-02-28 Classification: DNA BINDING PROTEIN Ligands: BTB |
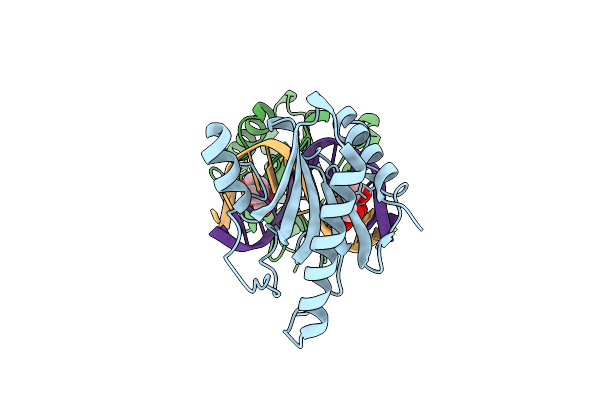 |
Structure Of Dna Binding Domain Of Mcrbc Endonuclease Bound To Dna: L68Y Mutant
Organism: Escherichia coli k-12, Dna molecule
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-02-28 Classification: DNA BINDING PROTEIN Ligands: BTB |
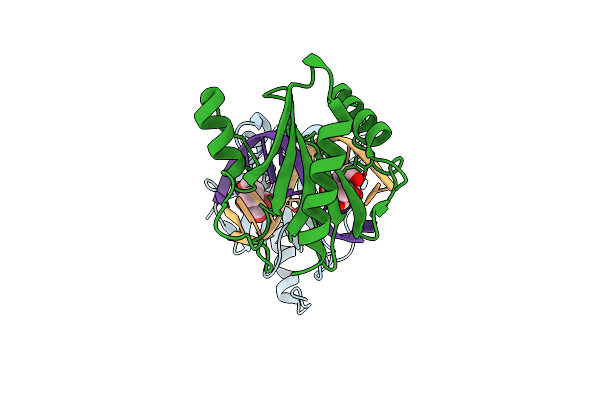 |
Structure Of Dna Binding Domain Of Mcrbc Endonuclease Bound To Hemimethylated Dna: L68F Mutant
Organism: Escherichia coli k-12, Dna molecule
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-02-28 Classification: DNA BINDING PROTEIN Ligands: BTB |
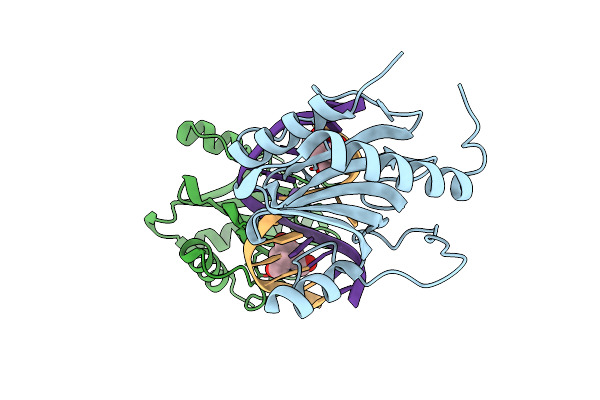 |
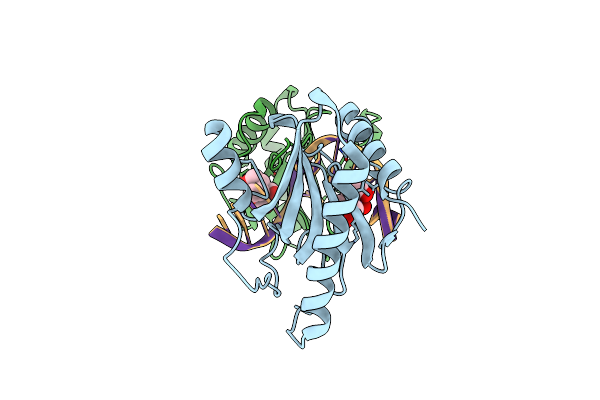
Structure Of Dna Binding Domain Of Mcrbc Endonuclease Bound To Dna: L68F Mutant
Organism: Escherichia coli k-12, Dna molecule
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-02-28 Classification: DNA BINDING PROTEIN Ligands: BTB |
 |
Structure Of Dna Binding Domain Of Mcrbc Endonuclease Bound To Dna: Y41F-L68Y Double Mutant
Organism: Escherichia coli k-12, Dna molecule
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-02-28 Classification: DNA BINDING PROTEIN Ligands: BTB |
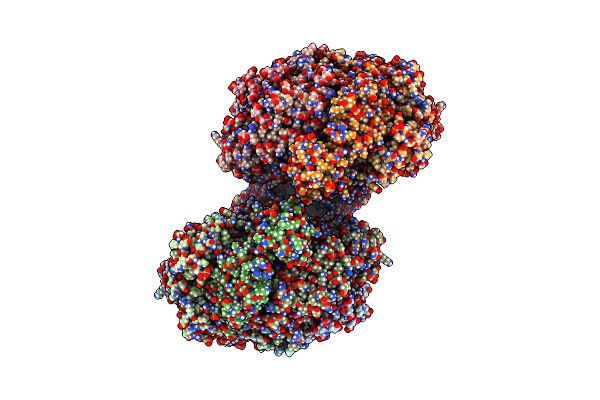 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2023-02-15 Classification: DNA BINDING PROTEIN Ligands: GNP, MG |
 |
Structure Of Mod Subunit Of The Type Iii Restriction-Modification Enzyme Mbo45V
Organism: Mycoplasma bovis
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2022-01-05 Classification: DNA BINDING PROTEIN Ligands: SFG, EDO |
 |
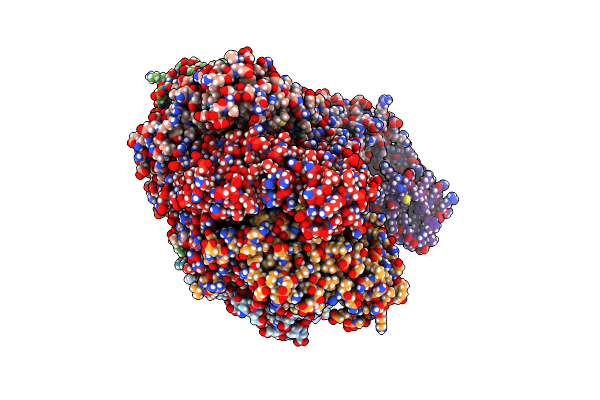
Organism: Staphylococcus aureus subsp. aureus usa300
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2021-01-20 Classification: DNA BINDING PROTEIN Ligands: SO4 |
 |
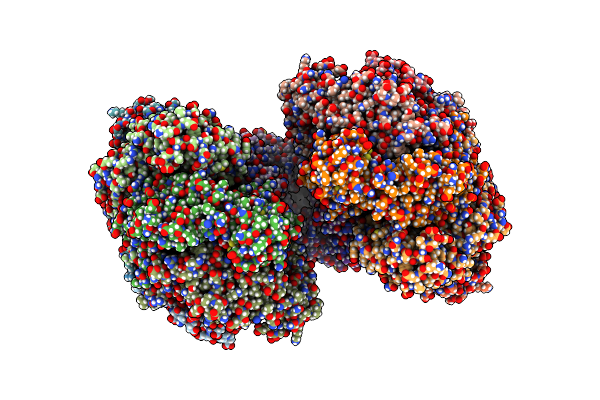
Structure Of Mcrbc Without Dna Binding Domains (One Half Of The Full Complex)
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2019-07-24 Classification: DNA BINDING PROTEIN Ligands: GNP, MG, GDP |
 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2019-07-24 Classification: DNA BINDING PROTEIN Ligands: GNP, MG, GDP |
 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2019-07-24 Classification: DNA BINDING PROTEIN Ligands: GNP, MG, GDP |
 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2019-07-24 Classification: DNA BINDING PROTEIN Ligands: GNP, MG, GDP |
 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2019-07-24 Classification: DNA BINDING PROTEIN Ligands: GNP, MG, GDP |
 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2019-07-24 Classification: DNA BINDING PROTEIN Ligands: GNP, MG, GDP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-05-15 Classification: DNA Ligands: K |
 |
Structure Of Human Telomeric Dna With 5-Selenophene-Modified Deoxyuridine At Residue 11
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-05-15 Classification: DNA Ligands: K |
 |
Structure Of Human Telomeric Dna With 5-Selenophene-Modified Deoxyuridine At Residue 12
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-05-15 Classification: DNA Ligands: K |
 |
Structure Of A Nuclease-Deletion Mutant Of The Type Isp Restriction-Modification Enzyme Llagi In Complex With A Dna Substrate Mimic
Organism: Lactococcus lactis, Unidentified
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2016-03-30 Classification: DNA BINDING PROTEIN/DNA |
 |
Atp-Dependent Type Isp Restriction-Modification Enzyme Llabiii Bound To Dna
Organism: Lactococcus lactis subsp. cremoris, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-09-16 Classification: HYDROLASE/DNA Ligands: K |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2012-03-21 Classification: HYDROLASE/DNA Ligands: SF4 |

