Search Count: 18
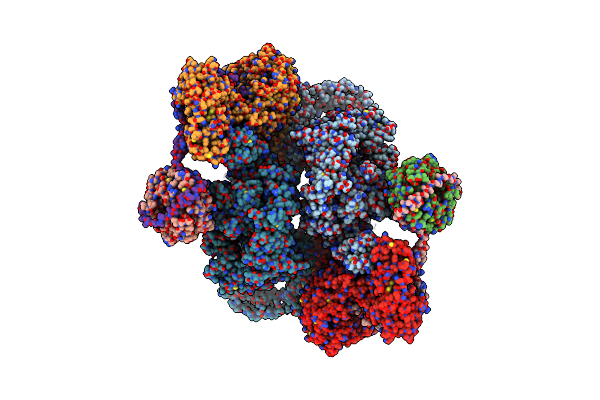 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-01-11 Classification: SIGNALING PROTEIN |
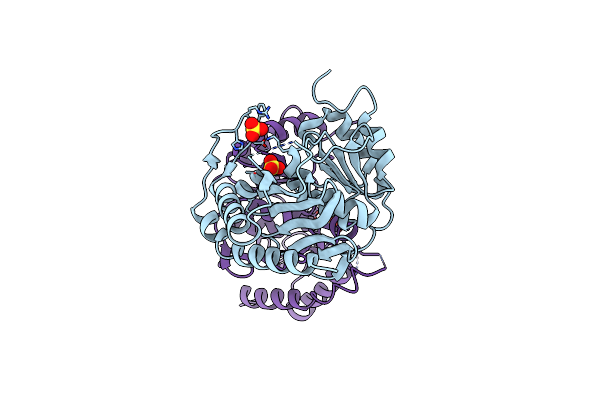 |
Crystal Strucuture Of An Inositol Monophosphatase Family Protein (Sas2203) From Staphylococcus Aureus Mssa476
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-01-18 Classification: HYDROLASE Ligands: SO4 |
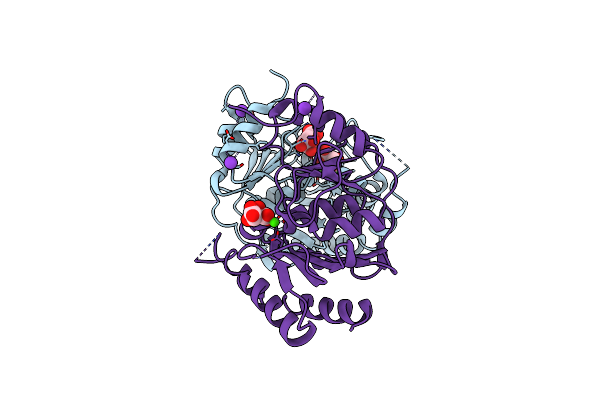 |
Crystal Structure Of Ca Bound Impase Family Protein From Staphylococcus Aureus
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2012-01-18 Classification: HYDROLASE Ligands: CA, SRT, K |
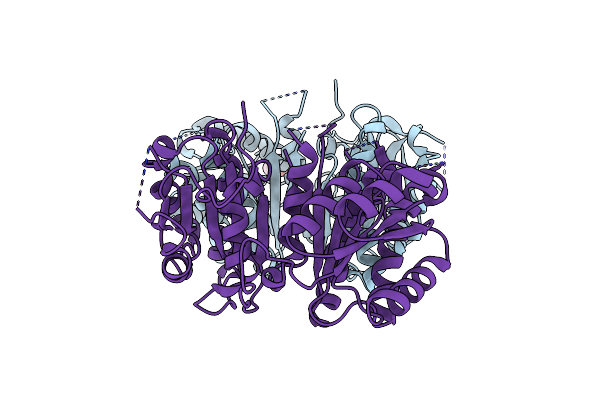 |
Crystal Structure Of A C-Terminal Trunacted Mutant Of A Putative Ketoacyl Reductase (Fabg4) From Mycobacterium Tuberculosis H37Rv At 2.5 Angstrom Resolution
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2010-12-22 Classification: OXIDOREDUCTASE Ligands: ACT |
 |
Crystal Structure Of Phosphate Bound Holo Glyceraldehyde-3-Phosphate Dehydrogenase 1 From Mrsa252 At 2.5 Angstrom Resolution
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-08-18 Classification: OXIDOREDUCTASE Ligands: NAD, PO4 |
 |
Crystal Structure Of C151G Mutant Of Glyceraldehyde 3-Phosphate Dehydrogenase 1 From Methicillin Resistant Staphylococcus Aureus (Mrsa252) At 2.5 Angstrom Resolution
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-08-18 Classification: OXIDOREDUCTASE Ligands: NAD, GOL, CL |
 |
Crystal Structure Of C151S+H178N Mutant Of Glyceraldehyde-3-Phosphate Dehydrogenase 1 (Gapdh1) From Staphylococcus Aureus Mrsa252 Complexed With Nad At 2.2 Angstrom Resolution
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-08-18 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Crystal Structure Of C151S+H178N Mutant Of Glyceraldehyde-3-Phosphate-Dehydrogenase 1 (Gapdh 1) From Staphylococcus Aureus Mrsa252 Complexed With Nad And G3P
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2010-08-18 Classification: OXIDOREDUCTASE Ligands: NAD, 3PG |
 |
Crystal Structure Of C151S Mutant Of Glyceraldehyde-3-Phosphate Dehydrogenase 1 (Gapdh 1)From Methicillin Resistant Staphylococcus Aureus Mrsa252 Complexed With Nad And G3P
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-08-18 Classification: OXIDOREDUCTASE Ligands: NAD, 3PG |
 |
Crystal Structure Of C151G Mutant Of Glyceraldehyde 3-Phosphate Dehydrogenase 1 (Gapdh1) From Methicillin Resistant Staphylococcus Aureus Mrsa252 Complexed With Nad And G3P
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-08-18 Classification: OXIDOREDUCTASE Ligands: 3PG, NAD |
 |
Crystal Structure Of Phosphate Bound Apo Glyceraldehyde-3-Phosphate Dehydrogenase 1 From Mrsa252 At 2.2 Angstrom Resolution
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-08-18 Classification: OXIDOREDUCTASE Ligands: PO4 |
 |
Crystal Structure Of H178N Mutant Of Glyceraldehyde-3-Phosphate-Dehydrogenase 1 (Gapdh 1) From Staphylococcus Aureus Mrsa252 Complexed With Nad At 2.0 Angstrom Resolution.
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-08-18 Classification: OXIDOREDUCTASE Ligands: NAD, GOL |
 |
Crystal Structure Of Thioacyl-Glyceraldehyde-3-Phosphate Dehydrogenase 1(Gapdh 1) From Methicillin Resistant Staphylococcus Aureus Mrsa252
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2010-08-18 Classification: OXIDOREDUCTASE Ligands: G3H, CL, GOL |
 |
Crystal Structure Of Apo Glyceraldehyde-3-Phosphate Dehydrogenase 1 (Gapdh1) From Methicllin Resistant Staphylococcus Aureus (Mrsa252)
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-08-18 Classification: OXIDOREDUCTASE Ligands: GOL |
 |
Crystal Structure Of Holo Glyceraldehyde-3-Phosphate Dehydrogenase 1 (Gapdh1) From Methicillin Resistant Staphylococcus Aureus Mrsa252 At 1.7 Angstrom Resolution
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2010-08-18 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Crystal Structure Of C151S Mutant Of Glyceraldehyde-3-Phosphate Dehydrogenase 1 (Gapdh1) Complexed With Nad From Staphylococcus Aureus Mrsa252 At 2.2 Angstrom Resolution
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-06-23 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Crystal Structure Of Mycobacterium Tuberculosis Low Molecular Protein Tyrosine Phosphatase (Mptpa) At 1.9A Resolution
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2005-03-22 Classification: HYDROLASE Ligands: CL |
 |
Crystal Structure Of Mycobacterium Tuberculosis Low Molecular Weight Protein Tyrosine Phosphatase (Mptpa) At 2.5A Resolution With Glycerol In The Active Site
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2005-03-22 Classification: HYDROLASE Ligands: CL, GOL |

