Search Count: 23
 |
Crystal Structure Of Octaheme Nitrite Reductase From Trichlorobacter Ammonificans In Complex With Nitrite
Organism: Trichlorobacter ammonificans
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-03-27 Classification: OXIDOREDUCTASE Ligands: HEC, NO, NO2, CA |
 |
Crystal Structure Of Octaheme Nitrite Reductase From Trichlorobacter Ammonificans In Space Group P21
Organism: Trichlorobacter ammonificans
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2024-03-27 Classification: OXIDOREDUCTASE Ligands: HEC, CA, PO4 |
 |
Organism: Shewanella oneidensis mr-1
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-04-06 Classification: TRANSFERASE Ligands: SO4, GOL, URA |
 |
Organism: Shewanella oneidensis mr-1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-04-06 Classification: TRANSFERASE Ligands: GOL, SO4, LI |
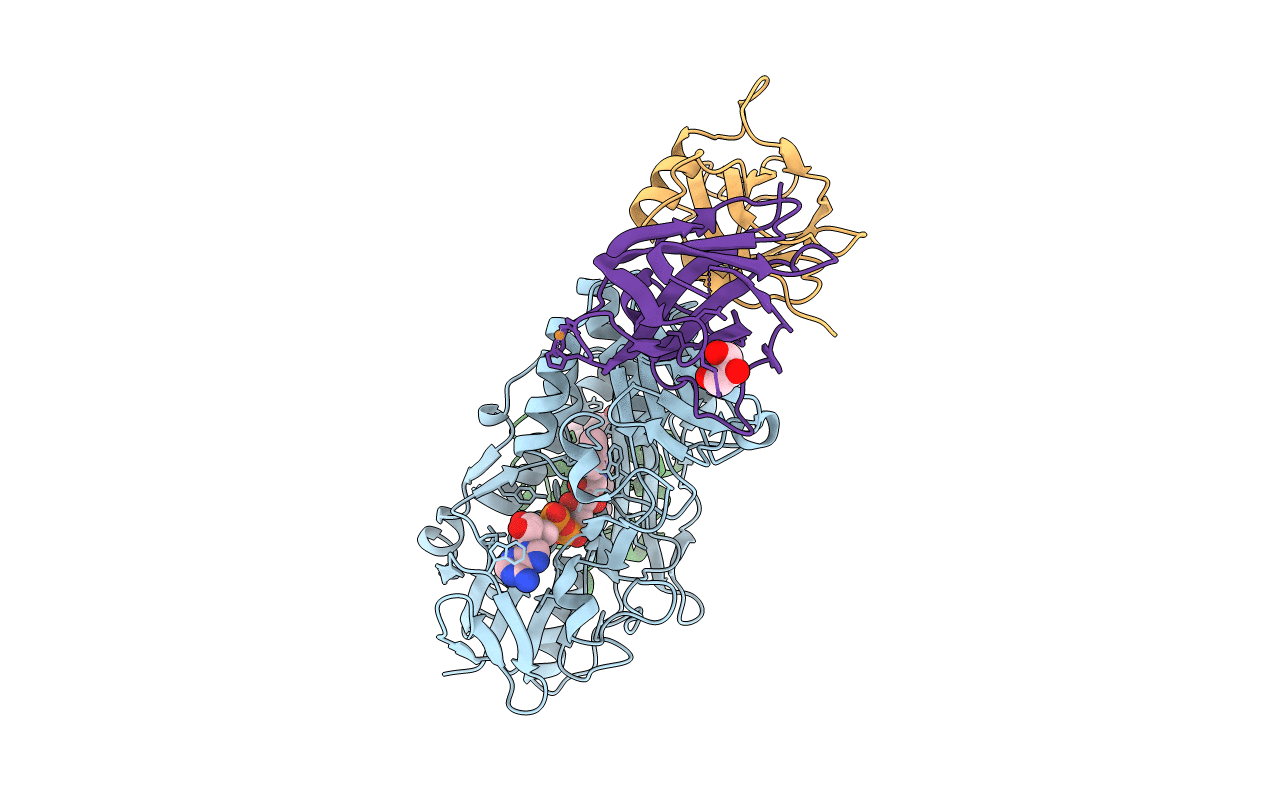 |
Crystal Structure Of Complex Between Flavocytochrome C And Copper Chaperone Copc From T. Paradoxus
Organism: Thioalkalivibrio paradoxus arh 1
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-02-28 Classification: OXIDOREDUCTASE Ligands: FAD, HEC, CU, GOL |
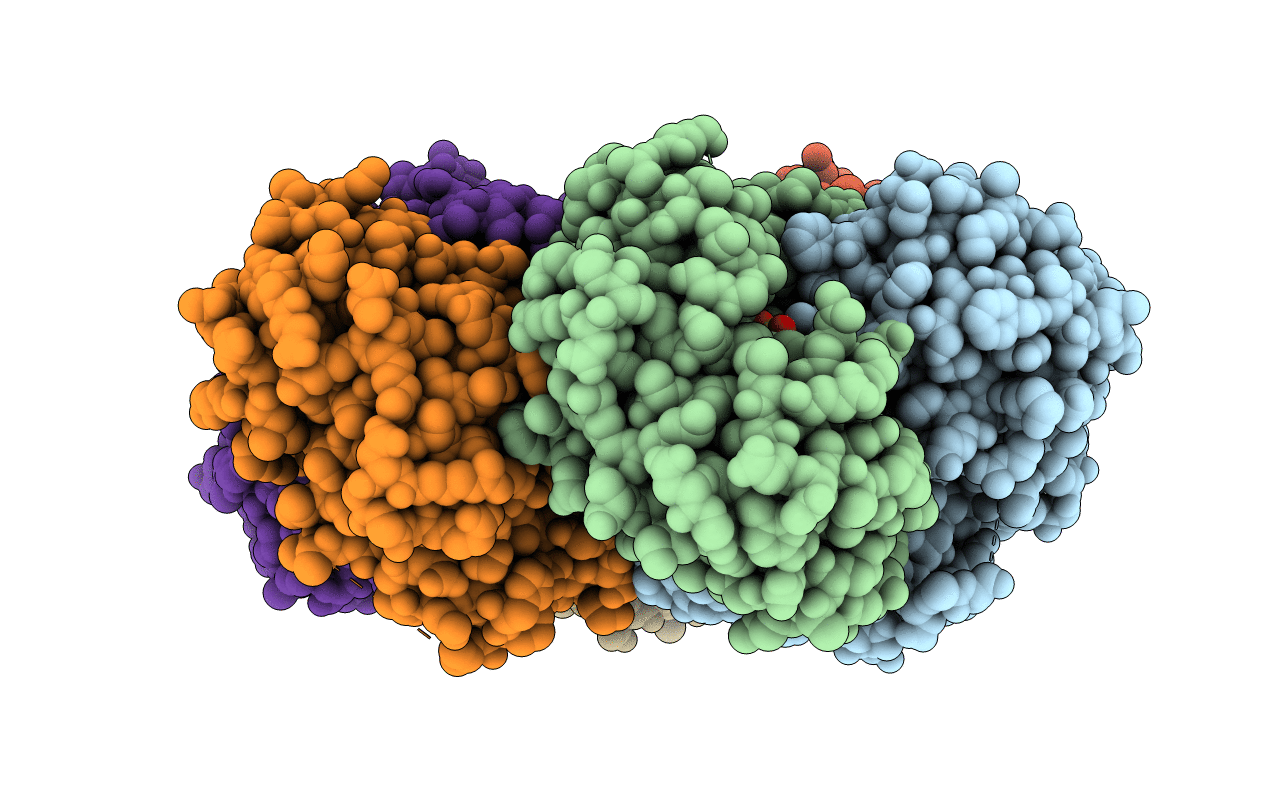 |
Crystal Structure Of C212S Mutant Of Shewanella Oneidensis Mr-1 Uridine Phosphorylase
Organism: Shewanella oneidensis (strain mr-1)
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2015-03-11 Classification: TRANSFERASE Ligands: URA, SO4 |
 |
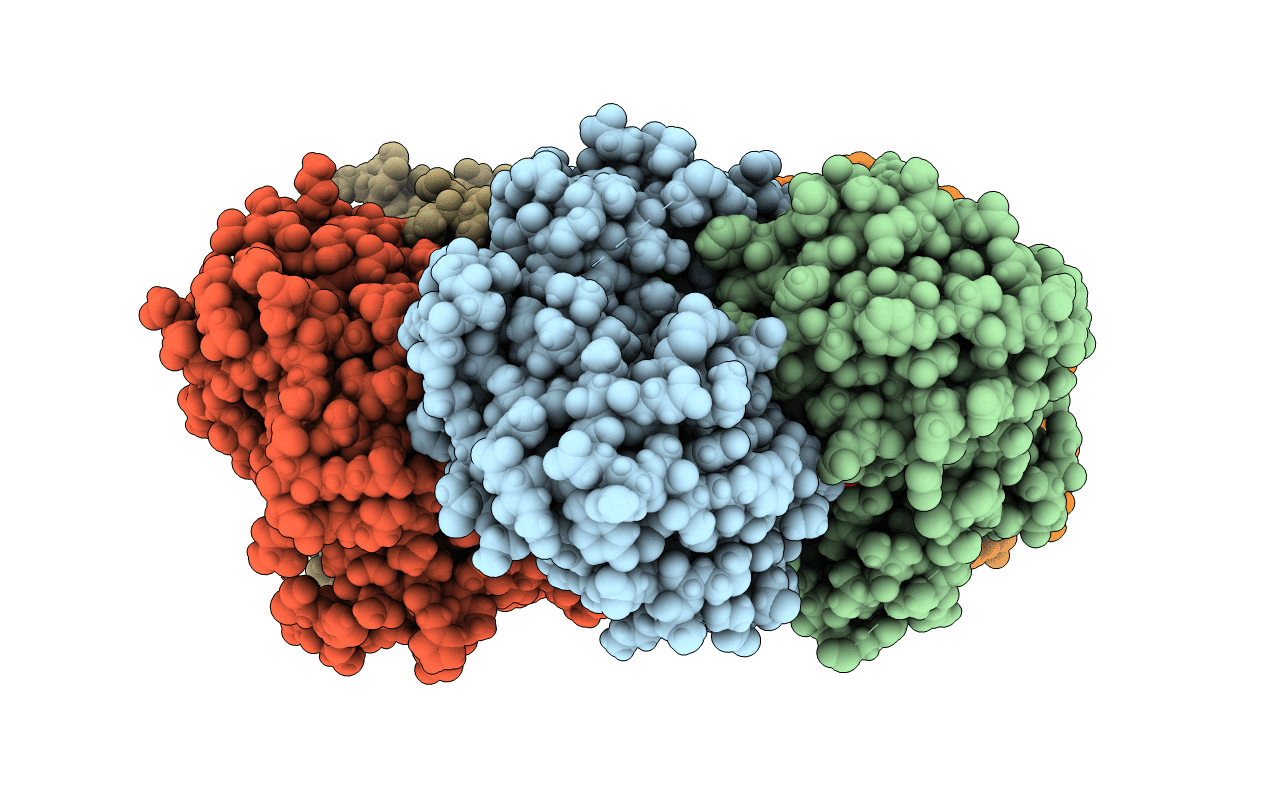
X-Ray Structure Of Uridine Phosphorylase From Shewanella Oneidensis Mr-1 In Complex With Uridine At 1.6 A Resolution
Organism: Shewanella oneidensis mr-1
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-12-10 Classification: TRANSFERASE Ligands: SO4, GOL, URI |
 |
Unique Conformation Of Uridine And Asymmetry Of The Hexameric Molecule Revealed In The High-Resolution Structures Of Shewanella Oneidensis Uridine Phosphorylase In The Free Form And In Complex With Uridine
Organism: Shewanella oneidensis
Method: X-RAY DIFFRACTION Resolution:0.93 Å Release Date: 2014-12-10 Classification: TRANSFERASE Ligands: SO4, CL, GOL |
 |
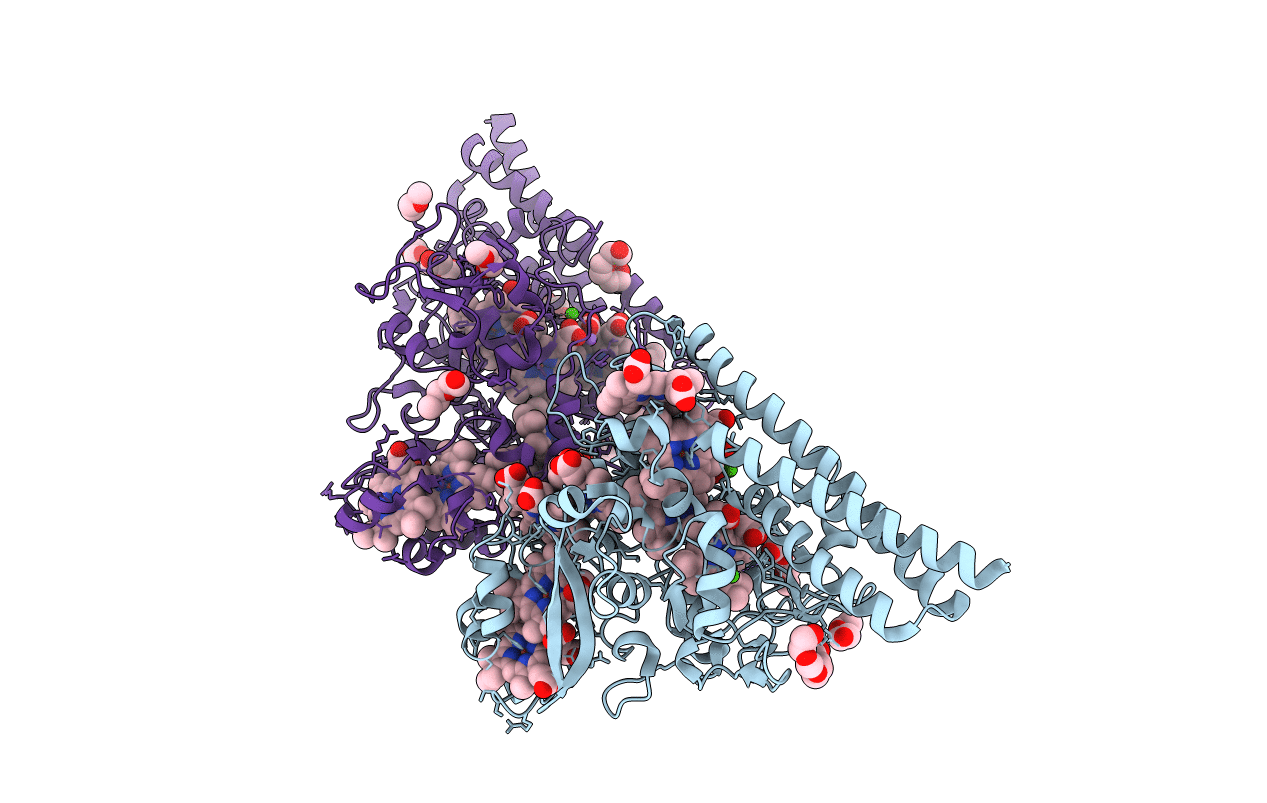
X-Ray Structure Of Iron Superoxide Dismutase From Acidilobus Saccharovorans
Organism: Acidilobus saccharovorans
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2012-06-27 Classification: OXIDOREDUCTASE Ligands: FE |
 |
Structure Of The Free Tvnirb Form Of Thioalkalivibrio Nitratireducens Cytochrome C Nitrite Reductase
Organism: Thioalkalivibrio nitratireducens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-01-25 Classification: OXIDOREDUCTASE Ligands: HEC, CA, PG4, NA |
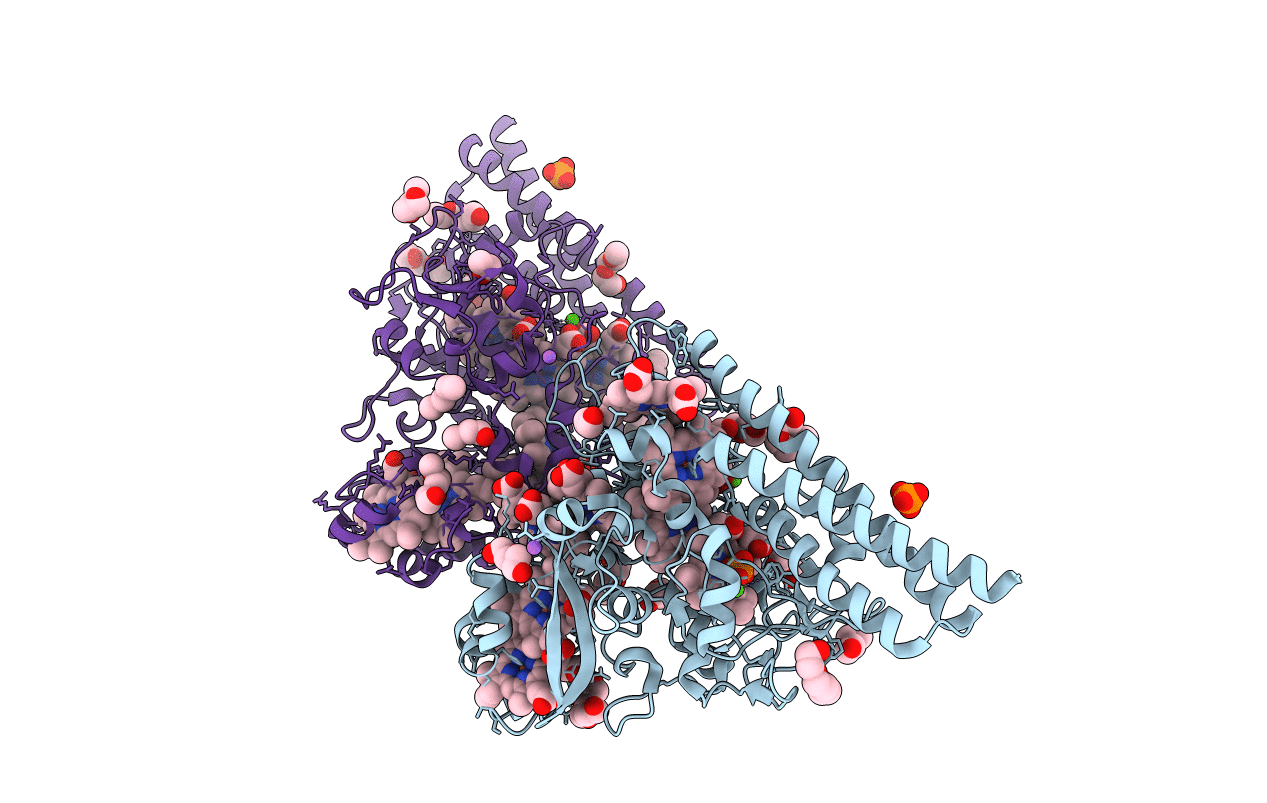 |
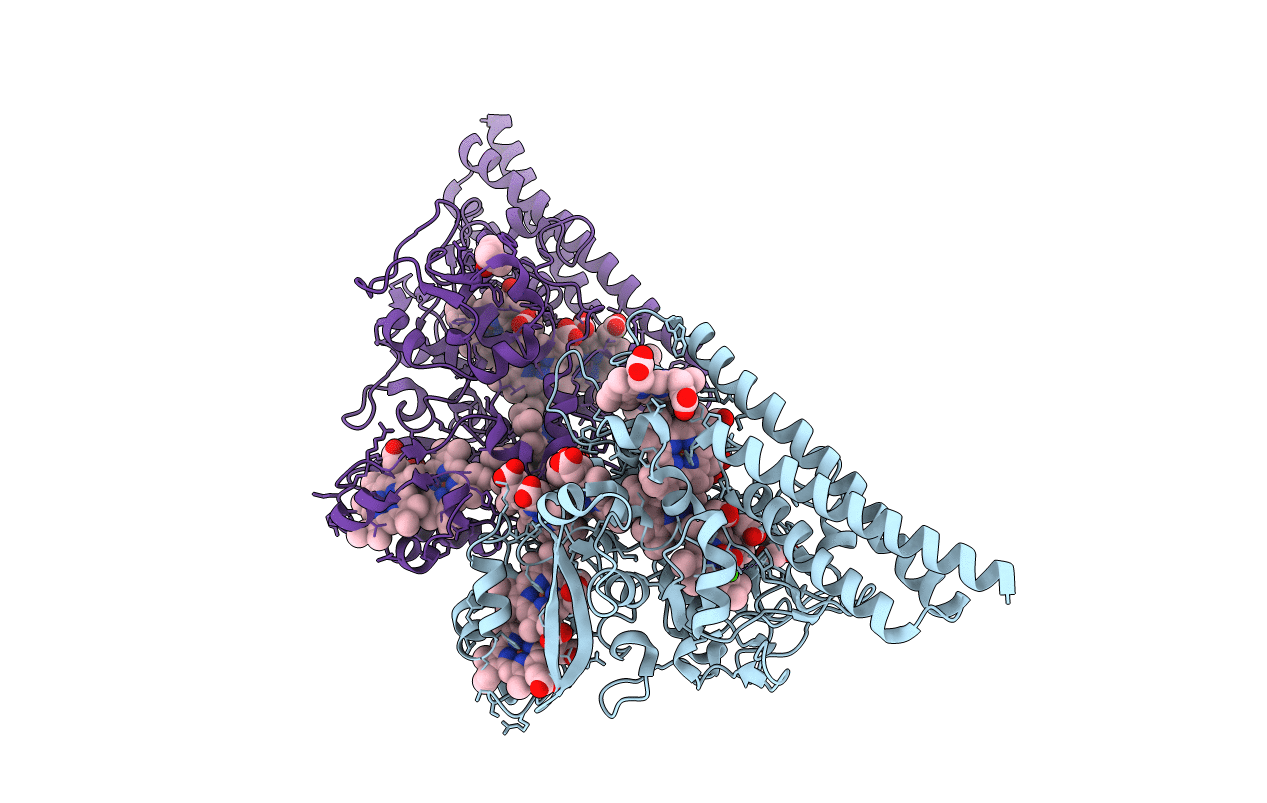
Structure Of The Thioalkalivibrio Nitratireducens Cytochrome C Nitrite Reductase With A Covalent Bond Between The Ce1 Atom Of Tyr303 And The Cg Atom Of Gln360 (Tvnirb)
Organism: Thioalkalivibrio nitratireducens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2011-07-06 Classification: OXIDOREDUCTASE Ligands: HEC, PO4, CA, PG4, PG6, NA |
 |
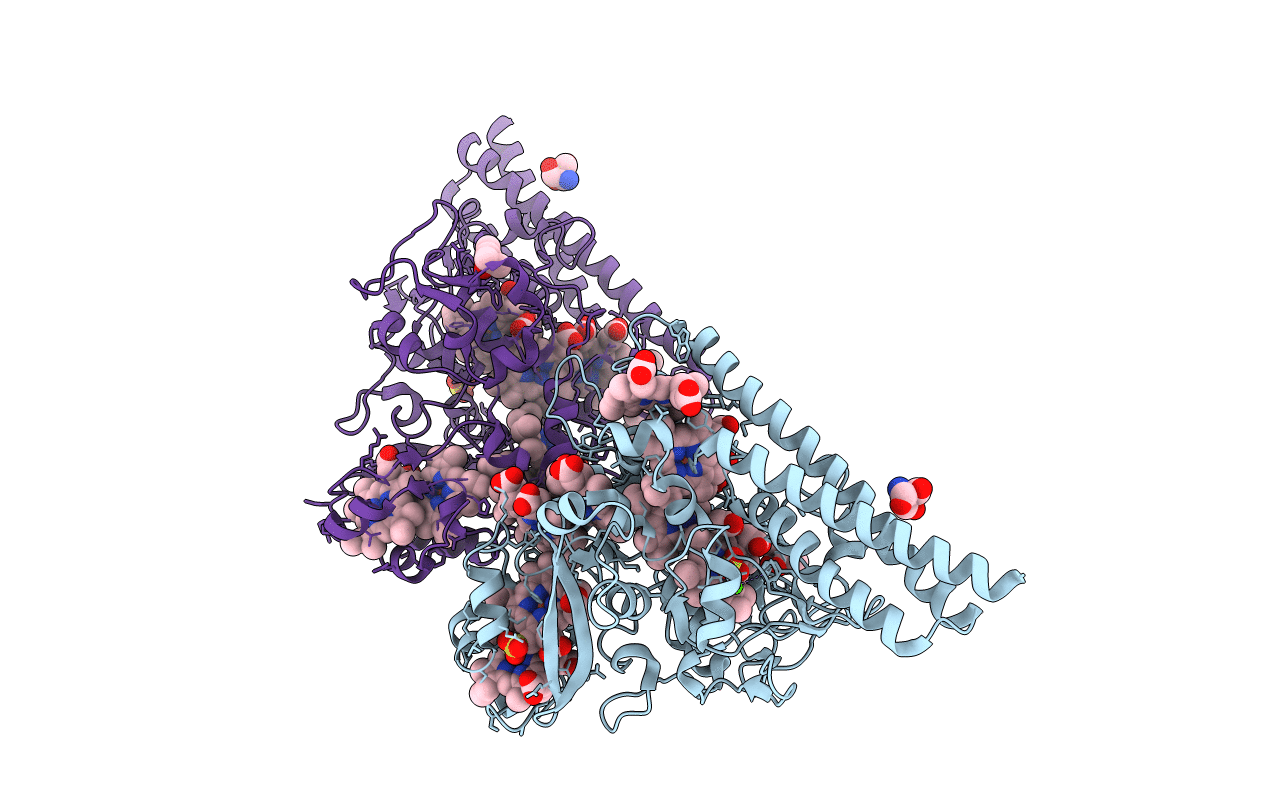
Structure Of The Thioalkalivibrio Nitratireducens Cytochrome C Nitrite Reductase In A Complex With Nitrite (Full Occupancy)
Organism: Thioalkalivibrio nitratireducens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2011-06-15 Classification: OXIDOREDUCTASE Ligands: HEC, NO2, CA, MRD |
 |
Structure Of The Thioalkalivibrio Nitratireducens Cytochrome C Nitrite Reductase In Complex With Sulfite (Modified Tyr-303)
Organism: Thioalkalivibrio nitratireducens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-02-16 Classification: OXIDOREDUCTASE Ligands: HEC, SO3, CA, MPD, TRS |
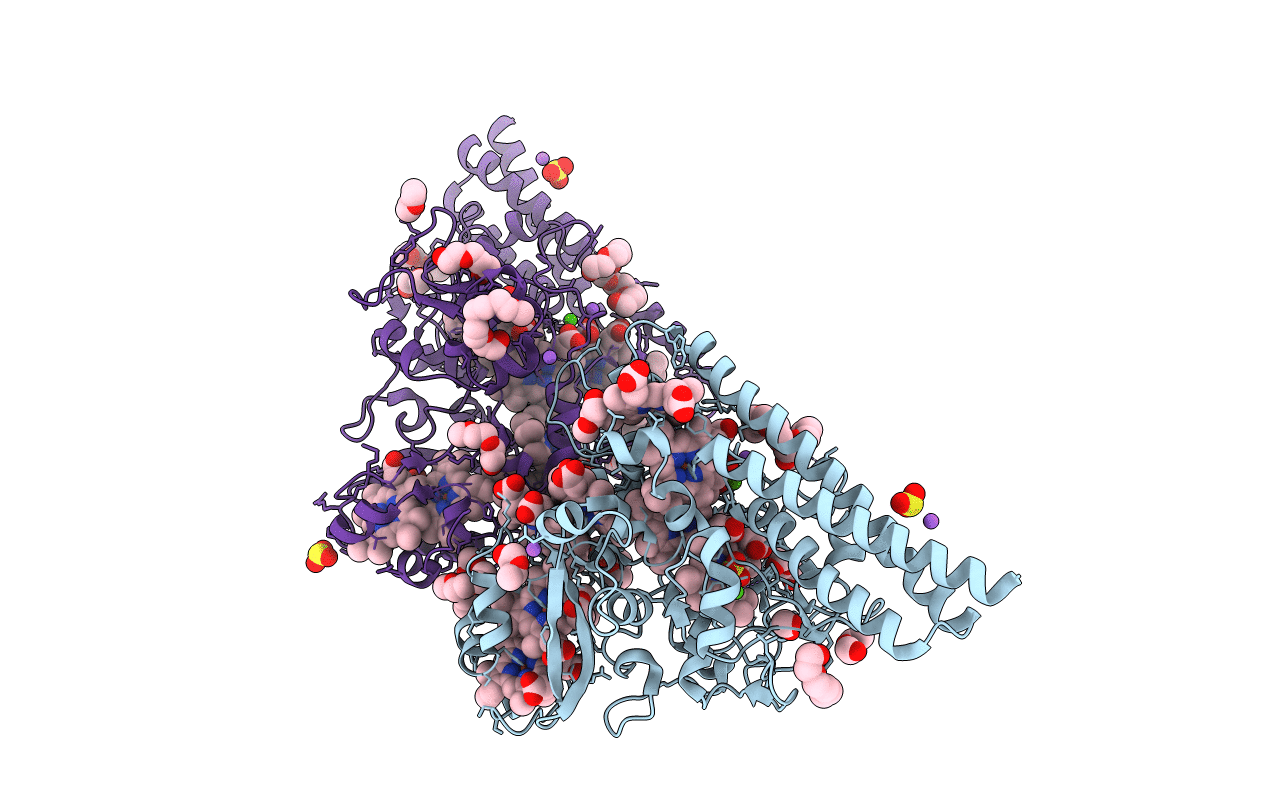 |
Structure Of The Thioalkalivibrio Nitratireducens Cytochrome C Nitrite Reductase Reduced By Sodium Borohydride (In Complex With Sulfite)
Organism: Thioalkalivibrio nitratireducens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2010-12-29 Classification: OXIDOREDUCTASE Ligands: HEC, SO3, CA, NA, PG6, PG4 |
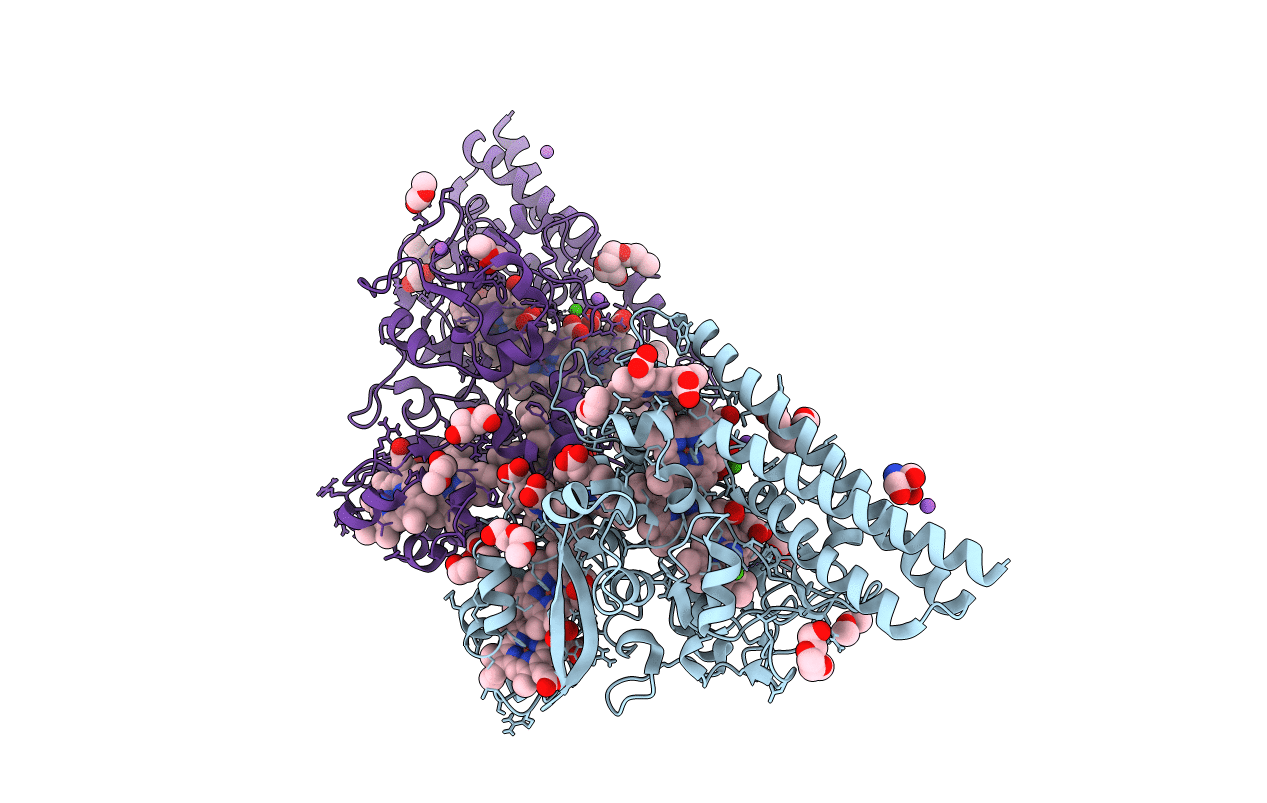 |
Structure Of The Thioalkalivibrio Nitratireducens Cytochrome C Nitrite Reductase In Complex With Cyanide
Organism: Thioalkalivibrio nitratireducens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2010-09-29 Classification: OXIDOREDUCTASE Ligands: HEC, CYN, CA, PG6, PG4, TRS, NA |
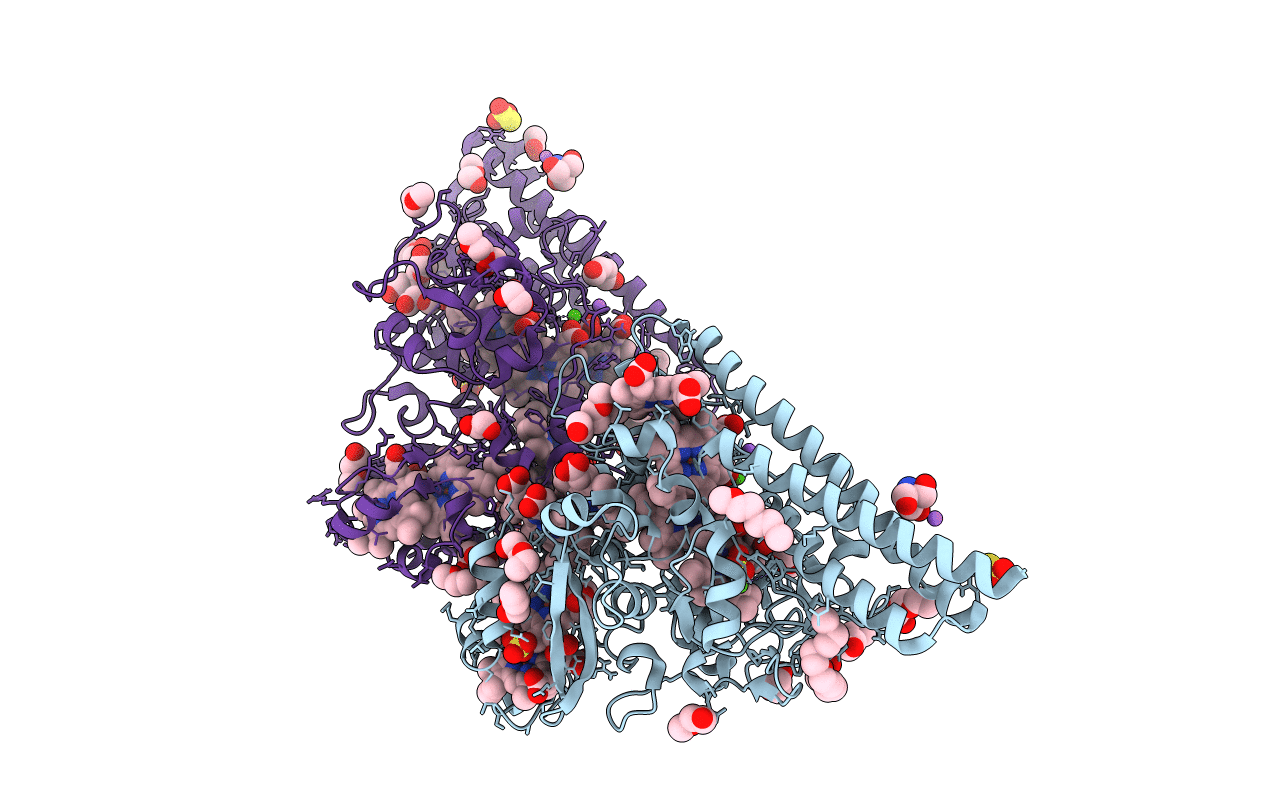 |
Structure Of The Thioalkalivibrio Nitratireducens Cytochrome C Nitrite Reductase Reduced By Sodium Dithionite (Sulfite Complex)
Organism: Thioalkalivibrio nitratireducens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2009-12-29 Classification: OXIDOREDUCTASE Ligands: HEC, SO3, CA, SO4, THJ, NA, PG6, TRS, PG4, ACT |
 |
Apo-Form Of Nad-Dependent Formate Dehydrogenase From Bacterium Moraxella Sp.C-1 In Closed Conformation
Organism: Moraxella sp.
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2009-12-01 Classification: OXIDOREDUCTASE Ligands: GOL, SO4 |
 |
Nad-Dependent Formate Dehydrogenase From Bacterium Moraxella Sp.C2 In Complex With Nad And Azide
Organism: Moraxella sp.
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2006-05-09 Classification: OXIDOREDUCTASE Ligands: AZI, NAD |
 |
Organism: Lupinus luteus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1995-10-15 Classification: OXYGEN TRANSPORT Ligands: HEM, OXY |
 |
Crystal Structure Of Ferric Complexes Of The Yellow Lupin Leghemoglobin With Isoquinoline At 1.8 Angstroms Resolution (Russian)
Organism: Lupinus luteus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1995-02-27 Classification: OXYGEN TRANSPORT Ligands: HEM, CMO |

