Search Count: 14
 |
Crystal Structure Of Udp-N-Acetylenolpyruvoylglucosamine Reductase (Murb) From Brucella Ovis
Organism: Brucella ovis atcc 25840
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2025-01-22 Classification: OXIDOREDUCTASE Ligands: SO4, FAD, EPU |
 |
Crystal Structure Of Cyclophilin Tgcyp23 From Toxoplasma Gondii In Complex With Nim811 (N-Methyl-4-Isoleucine Cyclosporin)
Organism: Toxoplasma gondii, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2024-10-30 Classification: ISOMERASE |
 |
Crystal Structure Of Cyclophilin Tgcyp23 From Toxoplasma Gondii In Complex With Alisporivir (Nonimmunosuppressive Analogue Of Cyclosporin)
Organism: Toxoplasma gondii, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2024-10-30 Classification: ISOMERASE |
 |
Crystal Structure Of Cyclophilin Tgcyp23 From Toxoplasma Gondii In Complex With Dihydro Cyclosporin A
Organism: Toxoplasma gondii, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2024-10-30 Classification: ISOMERASE |
 |
Crystal Structure Of A Dye-Decolorizing (Dyp) Peroxidase From Acinetobacter Radioresistens
Organism: Acinetobacter radioresistens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-01-25 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-01-20 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Crystal Structure Of Asp251Gly/Gln307His Mutant Of Cytochrome P450 Bm3 In Complex With N-Palmitoylglycine
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2016-01-20 Classification: OXIDOREDUCTASE Ligands: HEM, 140 |
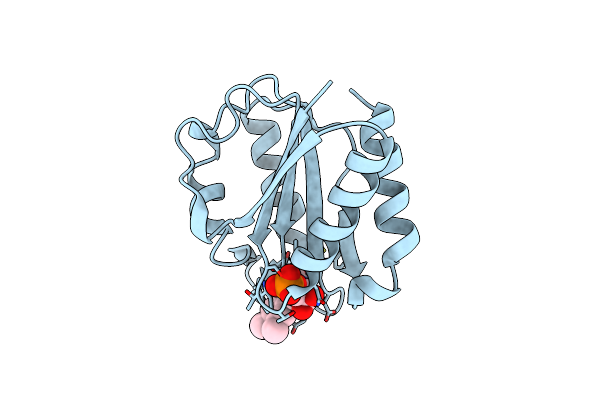 |
Low Temperature (100K) Crystal Structure Of Flavodoxin Mutant S64C, Dimer, Semiquinone State
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2004-12-07 Classification: ELECTRON TRANSPORT Ligands: FMN |
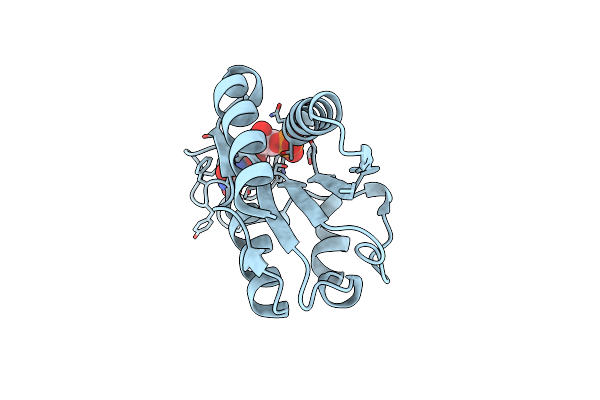 |
Low Temperature (100K) Crystal Structure Of Flavodoxin Mutant S64C, Monomer, Semiquinone State
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2004-12-07 Classification: ELECTRON TRANSPORT Ligands: FMN |
 |
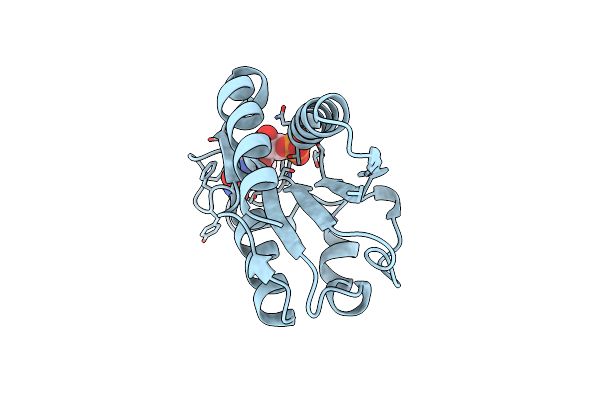
Low Temperature (100K) Crystal Structure Of Flavodoxin Mutant S64C, Homodimer, Oxidised State
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2004-12-07 Classification: ELECTRON TRANSPORT Ligands: FMN |
 |
Organism: desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2004-11-23 Classification: ELECTRON TRANSPORT Ligands: FMN |
 |
Low Temperature (100K) Crystal Structure Of Flavodoxin D. Vulgaris Wild-Type At 1.35 Angstrom Resolution
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2001-09-05 Classification: ELECTRON TRANSPORT Ligands: FMN |
 |
Low Temperature (100K) Crystal Structure Of Flavodoxin D. Vulgaris S35C Mutant At 1.44 Angstrom Resolution
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2001-09-05 Classification: ELECTRON TRANSPORT Ligands: FMN |
 |
Low Temperature (100K) Crystal Structure Of Flavodoxin D. Vulgaris S64C Mutant, Monomer Oxidised, At 2.4 Angstrom Resolution
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2001-09-05 Classification: ELECTRON TRANSPORT Ligands: FMN |

