Search Count: 928
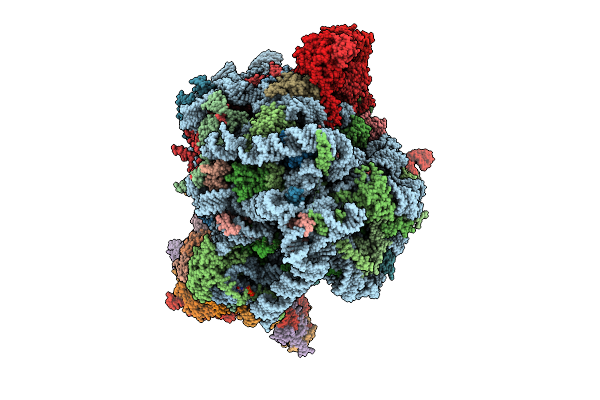 |
Human Quaternary Complex Of A Translating 80S Ribosome, Nac, Metap1 And Natd
Organism: Homo sapiens, Saccharomyces cerevisiae s288c, Aequorea victoria, Brachypodium distachyon
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: ZN, COA, GTP |
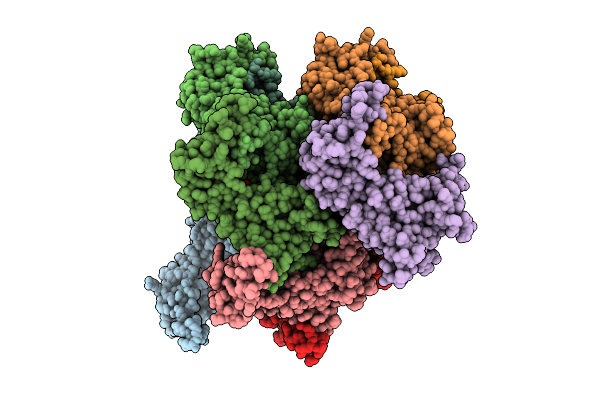 |
Eleven Polymer Msp1 From S.Cerevisiae (With A Catalytic Dead Mutaion) In Complex With An Unknown Peptide Substrate
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
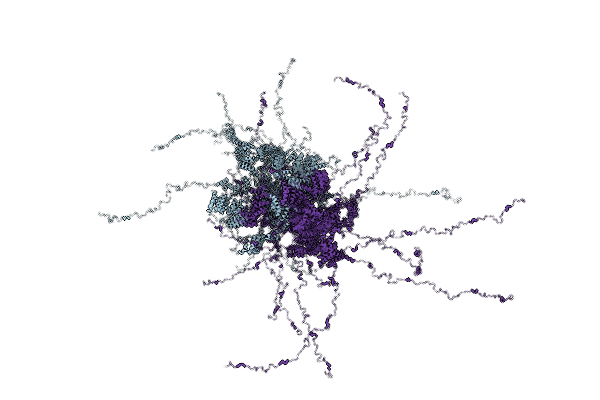 |
Organism: Saccharomyces cerevisiae s288c
Method: SOLUTION NMR Release Date: 2025-12-10 Classification: CHAPERONE |
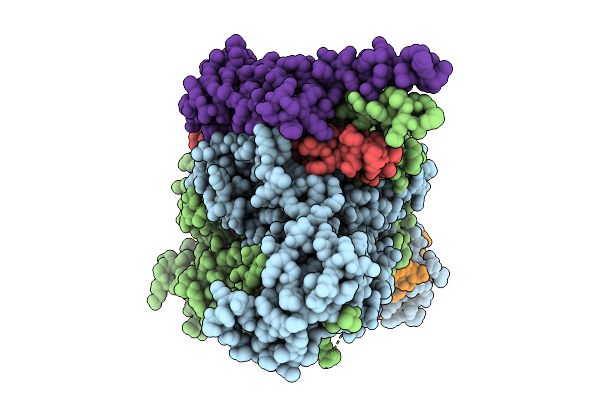 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: METAL BINDING PROTEIN Ligands: ZN |
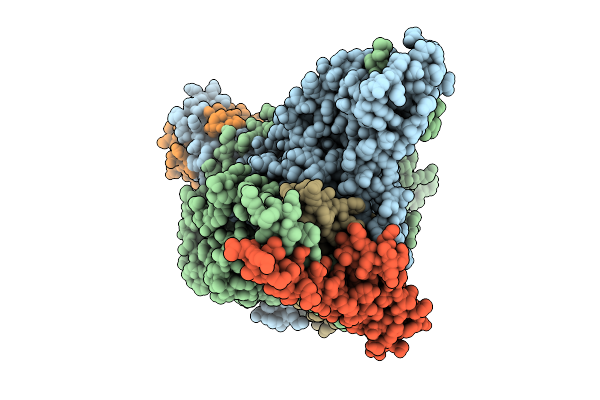 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: METAL BINDING PROTEIN Ligands: ZN, ACO |
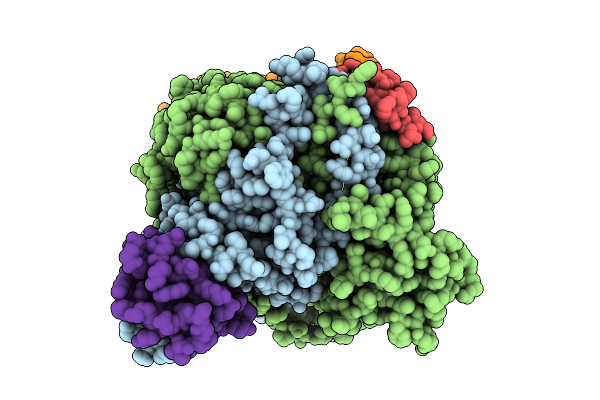 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSFERASE Ligands: ZN, ACO |
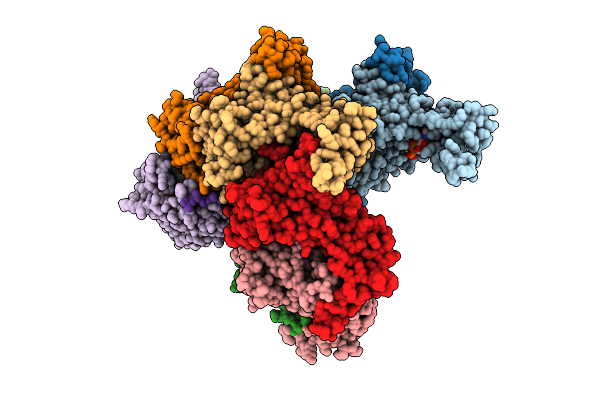 |
Nanomer Msp1 From S.Cerevisiae (With A Catalytic Dead Mutation) In Complex With An Unknown Peptide Substrate
Organism: Saccharomyces cerevisiae s288c, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
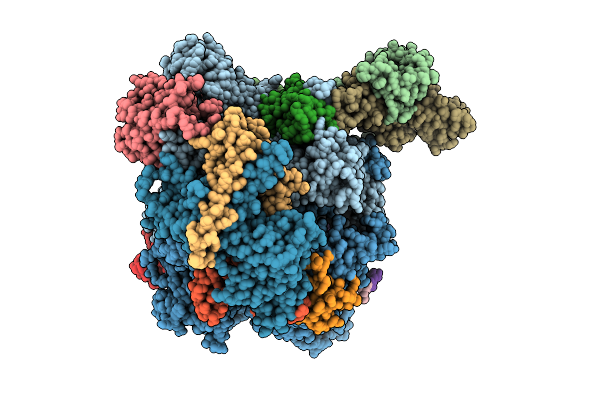 |
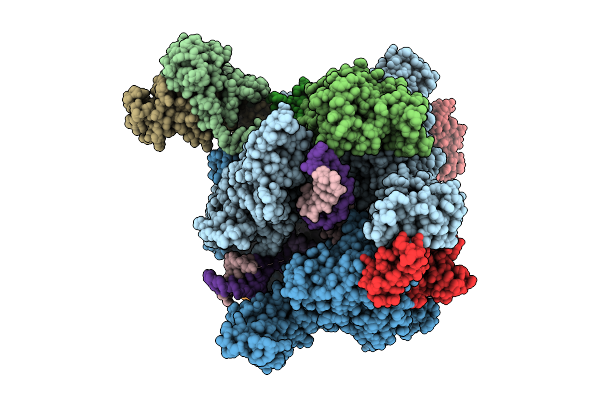
The Structure Of Rna Polymerase Ii Elongation Complex Paused At N-1 State By Actinomycin D.
Organism: Saccharomyces cerevisiae s288c, Synthetic construct, Streptomyces sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
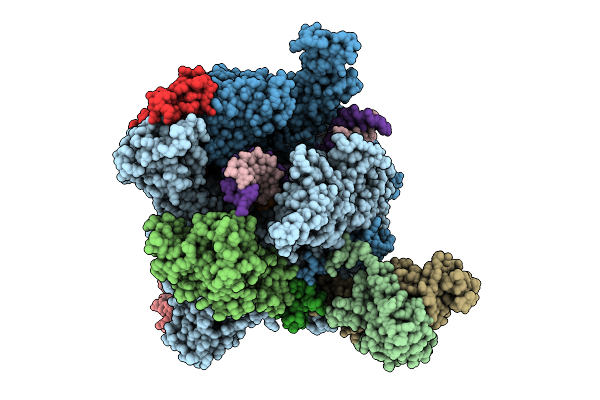
The Structure Of Rna Polymerase Ii Elongation Complex Paused At N-2 State By Actinomycin D.
Organism: Saccharomyces cerevisiae s288c, Synthetic construct, Streptomyces sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
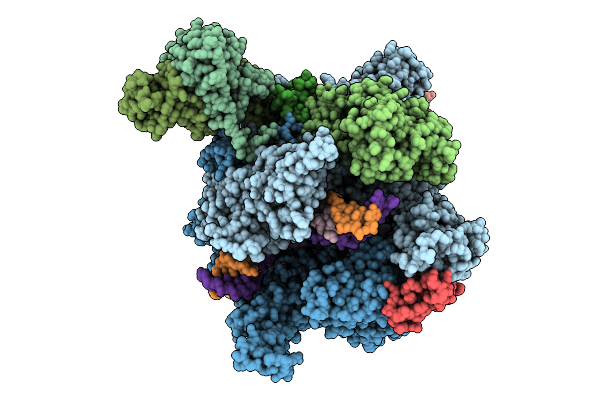
The Structure Of 1 Actd Bound To Rna Polymerase Ii Elongation Complex With 2 Ctg Repeats.
Organism: Synthetic construct, Saccharomyces cerevisiae s288c, Streptomyces sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
The Structure Of 2 Actd Bound To Rna Polymerase Ii Elongation Complex With 3 Ctg Repeats
Organism: Synthetic construct, Saccharomyces cerevisiae s288c, Streptomyces sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
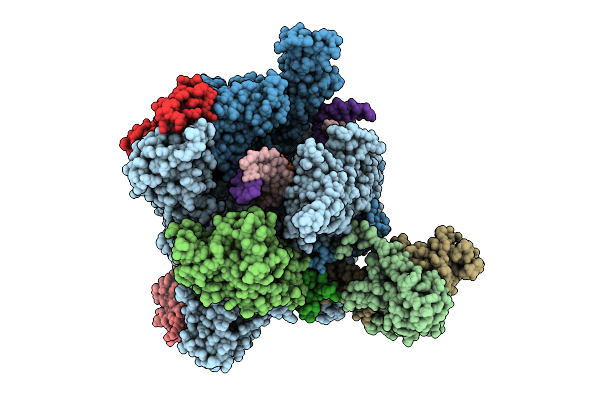
The Structure Of 3 Actd Bound To Rna Polymerase Ii Elongation Complex With 4 Ctg Repeats.
Organism: Synthetic construct, Saccharomyces cerevisiae s288c, Streptomyces sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
The Structure Of Rna Polymerase Ii Elongation Complex Paused At N-5 State By Actinomycin D.
Organism: Saccharomyces cerevisiae s288c, Synthetic construct, Streptomyces sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: ZN, MG |
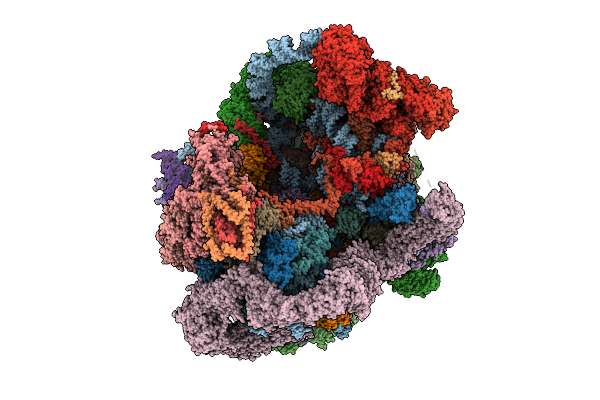 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME Ligands: MG, ATP, ZN, GTP |
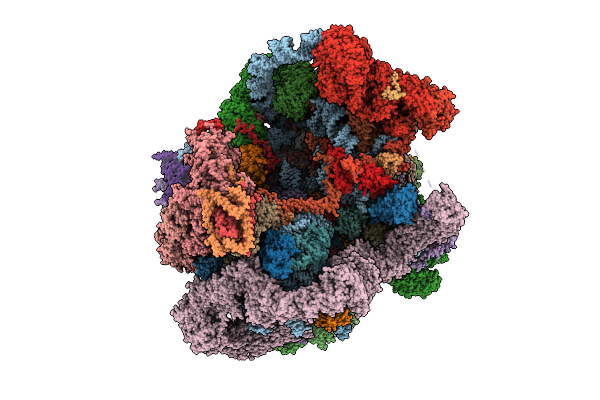 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME Ligands: MG, ATP, ZN, GTP |
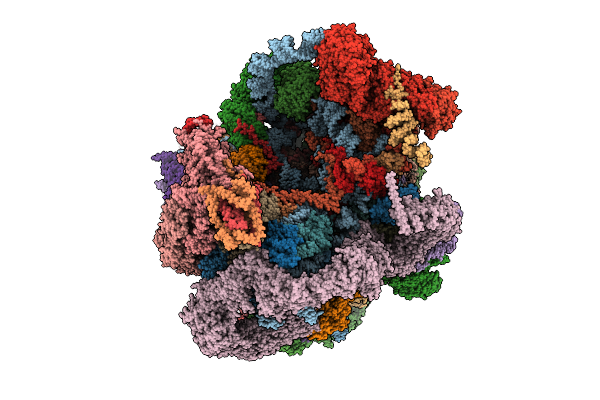 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME Ligands: MG, ATP, ZN, GTP |
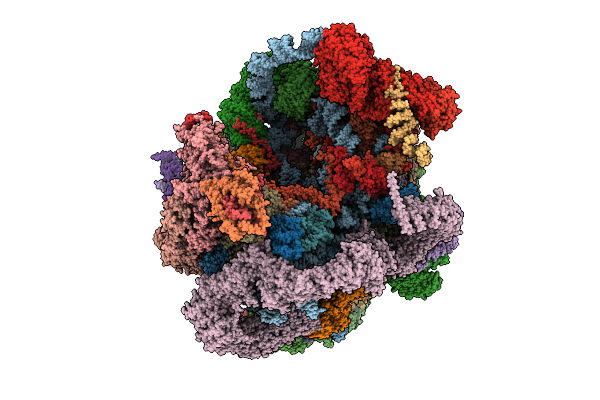 |
Organism: Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME Ligands: MG, ATP, ZN, GTP |
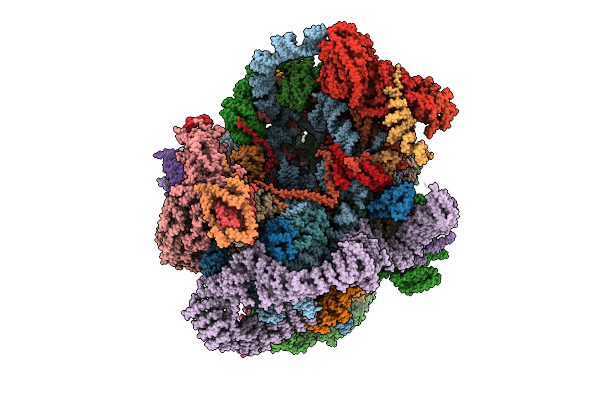 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME |
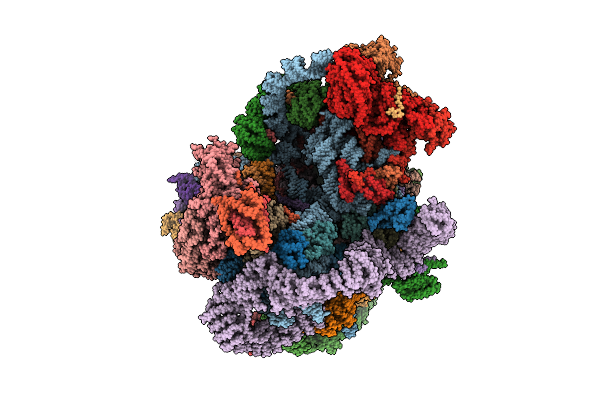 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME |
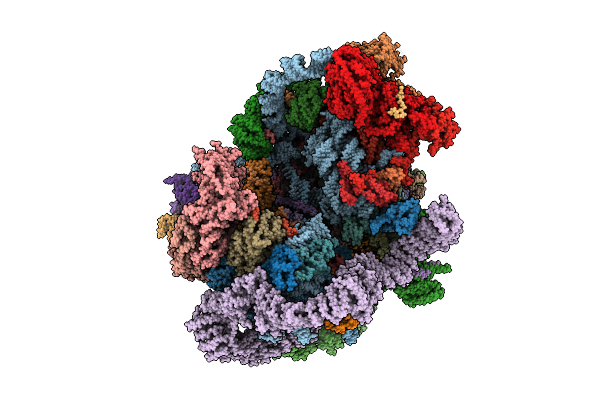 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME |

