Search Count: 1,080
 |
Organism: Acinetobacter baumannii 118362
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-09-25 Classification: TRANSFERASE Ligands: GOL, SO4 |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-09-20 Classification: STRUCTURAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2022-09-14 Classification: STRUCTURAL PROTEIN |
 |
Crystal Structure Of Sulfurtransferase (Dsrc Family Protein) From Acinetobacter Baumannii
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-12-08 Classification: TRANSFERASE |
 |
[4Fe-4S]-Dependent Thiouracil Desulfidase Tuds (Duf523Vcz) Solved By Fe-Sad Phasing
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2020-09-30 Classification: TRANSFERASE Ligands: SF4, EDO, PEG |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2020-09-30 Classification: TRANSFERASE Ligands: SF4, EDO, CL |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2020-09-30 Classification: TRANSFERASE Ligands: SF4, EDO, PGE, CL |
 |
[4Fe-4S]-Dependent Thiouracil Desulfidase Tuds (Duf523Vcz) Soaked With 4-Thiouracil (12.65 Kev, 7.125 Kev (Fe-Sad) And 6.5 Kev (S-Sad) Data)
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2020-09-30 Classification: TRANSFERASE Ligands: SF4, H2S, EDO, PEG |
 |
[4Fe-4S]-Dependent Thiouracil Desulfidase Tuds (Duf523Vcz) Soaked With 4-Thiouracil (S-Sad Data)
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2020-09-30 Classification: TRANSFERASE Ligands: SF4, EDO, H2S, PEG |
 |
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-09-09 Classification: TRANSFERASE Ligands: MG, CL, EDO, CA, AVJ, ZN |
 |
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2020-09-09 Classification: TRANSFERASE Ligands: AVJ, MG, CL |
 |
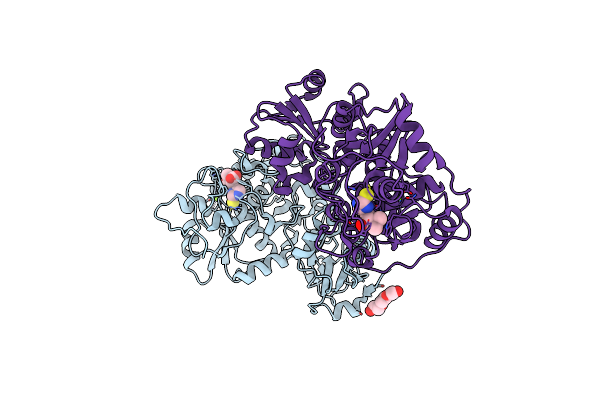
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2020-09-09 Classification: TRANSFERASE Ligands: AVJ, MG, CL, PEG |
 |
The Structure Of Eanb/Y353F-Cys412-Persulfide In Tetrahedral Intermediate State With Ergothioneine
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-09-09 Classification: TRANSFERASE Ligands: CL, MG, BR, PGE, LW8 |
 |
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-08-26 Classification: TRANSFERASE Ligands: AVJ, MG, CL, GOL, EDO, PEG, PG4, PDO, IMD, NI |
 |
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2020-08-26 Classification: TRANSFERASE Ligands: MG, AVJ, CL, PGE, GOL, IMD, NA, EDO |
 |
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2020-08-26 Classification: TRANSFERASE Ligands: MG, CL, PEG, MPO, EDO, PGE |
 |
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-08-26 Classification: TRANSFERASE Ligands: MG, AVJ, PEG, EDO, CL, BR |
 |
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-08-26 Classification: TRANSFERASE Ligands: MG, LW8, CL, EDO |
 |
The Structure Of Eanb/Y353A Complex With Ergothioneine Covalent Linked With Persulfide Cys412
Organism: Chlorobium limicola
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2020-08-26 Classification: TRANSFERASE Ligands: DV6, MG, CL, GOL, IMD, ZN, BR, PEG, EDO |
 |
Organism: Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-07-22 Classification: TRANSFERASE Ligands: ZN |

