Search Count: 476
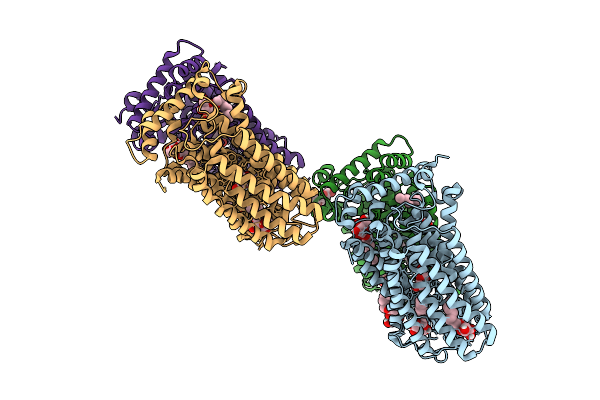 |
Structure Of The Malarial Hexose Transporter 1 (Pfht1) In Complex With 2,5-Anhydro-D-Mannitol.
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSPORT PROTEIN Ligands: BNG, A1IVP |
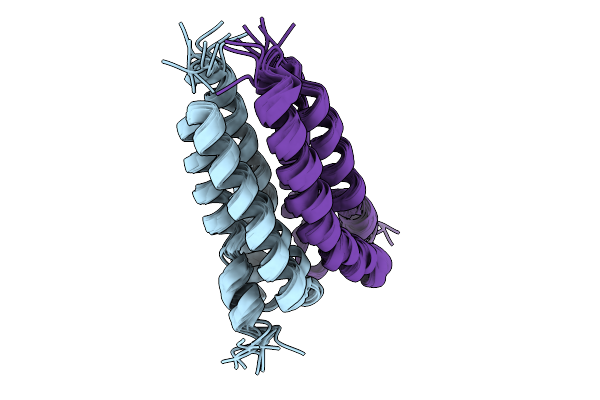 |
The Outward-Open Structure Of Bjsemisweet In Native Cellular Membranes Determined By In Situ Solid-State Nmr
Organism: Bradyrhizobium diazoefficiens usda 110
Method: SOLID-STATE NMR Release Date: 2025-09-10 Classification: MEMBRANE PROTEIN |
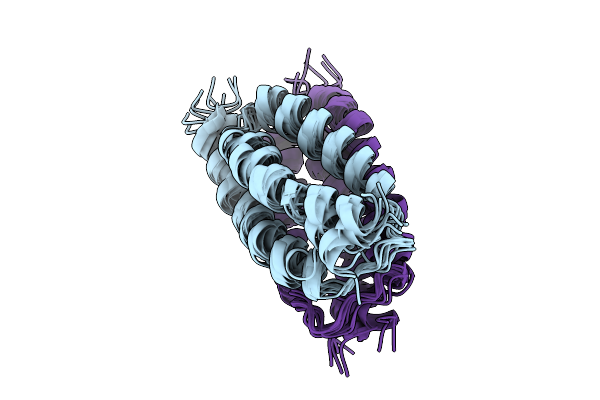 |
The Occluded Structures Of Bjsemisweet In Native Cellular Membranes Determined By In Situ Solid-State Nmr
Organism: Bradyrhizobium diazoefficiens usda 110
Method: SOLID-STATE NMR Release Date: 2025-09-10 Classification: MEMBRANE PROTEIN |
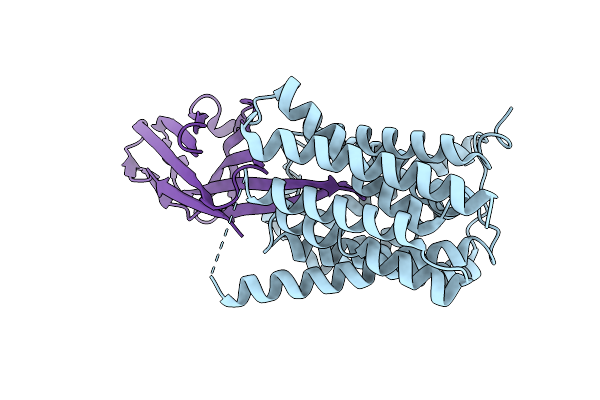 |
Cryo-Em Structure Of Candida Albicans Vrg4 Bound To An Inhibitory Nanobody.
Organism: Lama glama, Candida albicans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: TRANSPORT PROTEIN |
 |
Cryo-Em Structure Of Candida Albicans Vrg4 Bound To An Inhibitory Nanobody And Gdp-Mannose.
Organism: Candida albicans, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: TRANSPORT PROTEIN Ligands: GDD |
 |
Organism: Vibrio sp. (strain n418)
Method: SOLID-STATE NMR Release Date: 2024-12-04 Classification: MEMBRANE PROTEIN |
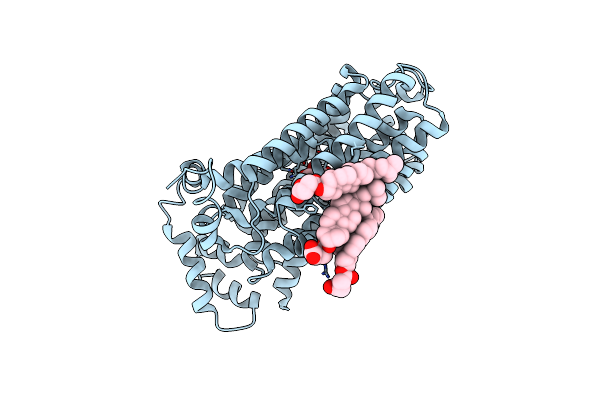 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2024-08-14 Classification: MEMBRANE PROTEIN Ligands: BGC, Y01 |
 |
Crystal Structure Of Sar11_0769 From 'Candidatus Pelagibacter Ubique' Htcc1062 Bound To A Co-Purified Ligand, Beta-Galactopyranose
Organism: Candidatus pelagibacter ubique htcc1062
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2024-07-17 Classification: TRANSPORT PROTEIN Ligands: GAL |
 |
Crystal Structure Of A Ligand-Bound Cation-Site Mutant D55C Of A Melibiose Transporter
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-06-05 Classification: MEMBRANE PROTEIN Ligands: 1PE, LMO |
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. lt2
Method: X-RAY DIFFRACTION Resolution:3.18 Å Release Date: 2024-06-05 Classification: MEMBRANE PROTEIN Ligands: 1PE |
 |
Cryoem Structure Of An Inward-Facing Melbst At A Na(+)-Bound And Sugar Low-Affinity Conformation
Organism: Salmonella enterica subsp. enterica serovar typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-02-28 Classification: TRANSPORT PROTEIN |
 |
Crystal Structure Of The Glucose-Binding Protein Sar11_0769 From "Candidatus Pelagibacter Ubique" Htcc1062 Bound To Glucose
Organism: Candidatus pelagibacter ubique htcc1062
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2023-12-27 Classification: TRANSPORT PROTEIN Ligands: BGC |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.30 Å Release Date: 2023-11-22 Classification: TRANSPORT PROTEIN Ligands: NAG, LN9 |
 |
Cryo-Em Structure Of The Tripartite Atp-Independent Periplasmic (Trap) Transporter Siaqm From Haemophilus Influenzae (Parallel Dimer)
Organism: Haemophilus influenzae rd kw20
Method: ELECTRON MICROSCOPY Release Date: 2023-11-22 Classification: TRANSPORT PROTEIN Ligands: PGT, NA |
 |
Cryo-Em Structure Of The Tripartite Atp-Independent Periplasmic (Trap) Transporter Siaqm From Haemophilus Influenzae (Antiparallel Dimer)
Organism: Haemophilus influenzae rd kw20
Method: ELECTRON MICROSCOPY Release Date: 2023-11-22 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.80 Å Release Date: 2023-11-15 Classification: TRANSPORT PROTEIN Ligands: NAG, LE6, NA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.10 Å Release Date: 2023-11-15 Classification: TRANSPORT PROTEIN Ligands: NAG, LFL, NA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.10 Å Release Date: 2023-11-08 Classification: TRANSPORT PROTEIN Ligands: NAG, L3R, NA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.90 Å Release Date: 2023-11-01 Classification: TRANSPORT PROTEIN Ligands: NAG, KZ3, NA |
 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-09-06 Classification: TRANSPORT PROTEIN Ligands: ATP |

