Search Count: 17,328
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: NUCLEAR PROTEIN Ligands: GOL, A1BI5, EDO |
 |
Crystal Structure Of Human Prostaglandin Reductase 1 (Ptgr1) In Complex With Nadp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: CL, NAP, PG6 |
 |
Crystal Structure Of Human Prostaglandin Reductase 1 (Ptgr1) In Complex With Nadp And Indomethacin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: ACT, IMN, NAP, GOL |
 |
Crystal Structure Of Human Prostaglandin Reductase 1 (Ptgr1) In Complex With Nadp And Indomethacin (Orthorhombic P Form)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: CL, SO4, GOL, IMN, NAP, PG4 |
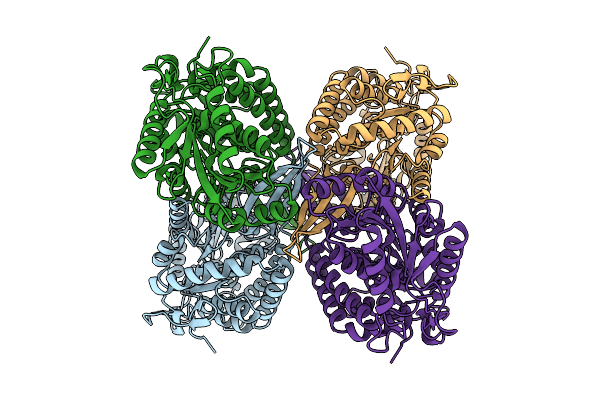 |
Cryoem Structure Of Aldehyde Dehydrogenase From Burkholderia Cenocepacia At 2.33A Resolution
Organism: Burkholderia cenocepacia
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: OXIDOREDUCTASE |
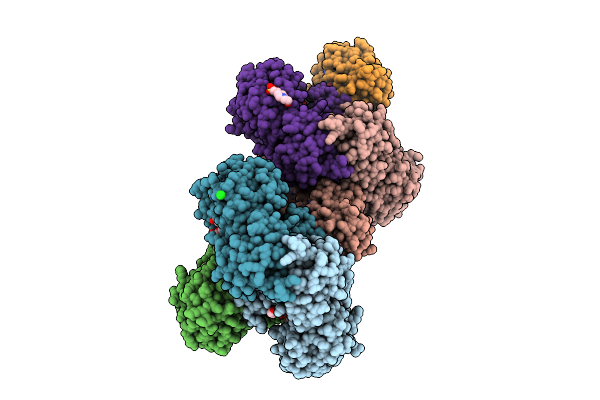 |
Crystal Structure Of A Glyceraldehyde-3-Phosphate Dehydrogenase From Neisseria Gonorrhoeae In Complex With Nad And Glyceraldehyde-3-Phosphate
Organism: Neisseria gonorrhoeae nccp11945
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: OXIDOREDUCTASE Ligands: G3H, PEG, NAD, NA, PGE, CL, GOL, EPE |
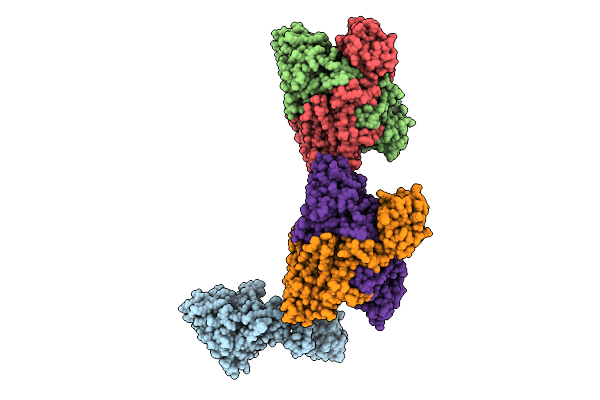 |
Crystal Structure Of Formyl-Coenzyme A Transferase From Brucella Melitensis In Complex With Succinate
Organism: Brucella melitensis 16m1w
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: TRANSFERASE Ligands: SIN |
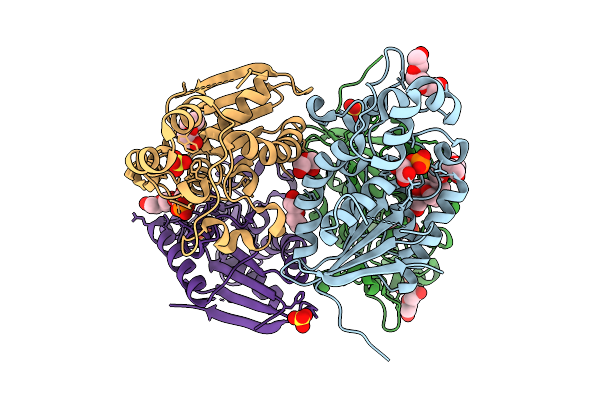 |
Crystal Structure Of Phosphoglycerate Mutase From Trichomonas Vaginalis In Complex With 3-Phosphoglyceric Acid
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: ISOMERASE Ligands: CL, 3PG, PG4, SO4 |
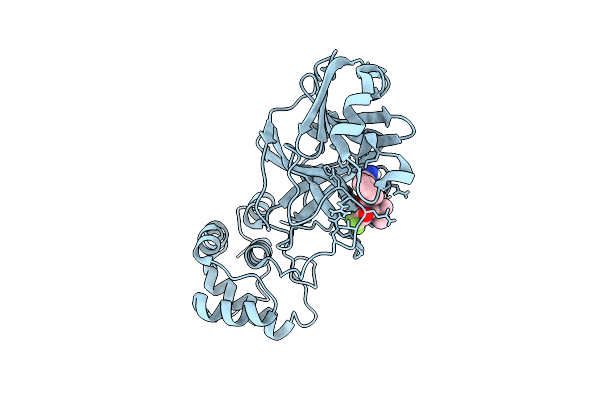 |
X-Ray Structure Of Sars-Cov-2 Main Protease V186G Covalently Bound To Compound Grl-051-22 At 1.3 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN, HYDROLASE Ligands: A1BFE |
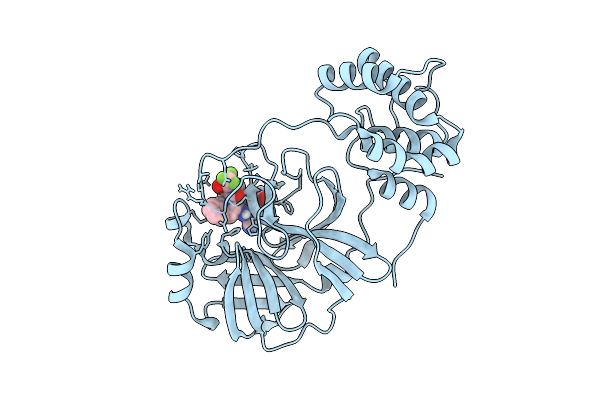 |
X-Ray Structure Of Sars-Cov-2 Main Protease Covalently Bound To Compound Grl-051-22 At 1.75 A.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN, HYDROLASE Ligands: A1BFE |
 |
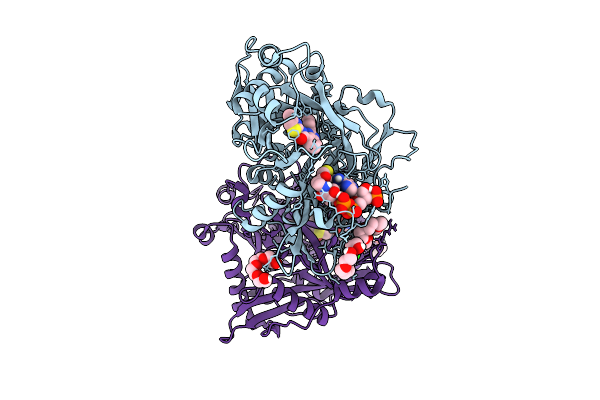
X-Ray Structure Of Sars-Cov-2 Main Protease T190I Covalently Bound To Compound Grl-051-22 At 1.5 A
Organism: Severe acute respiratory syndrome coronavirus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN, HYDROLASE Ligands: A1BFE, NA |
 |
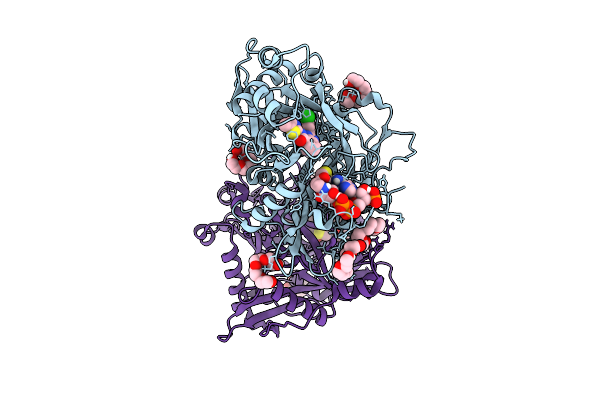
Crystal Structure Of Cryptosporidium Parvum N-Myristoyltransferase With Bound Myristoyl-Coa And Inhibitor 20045
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE/INHIBITOR Ligands: MYA, A1BHP, CL, PG4, 1PE, PGE |
 |
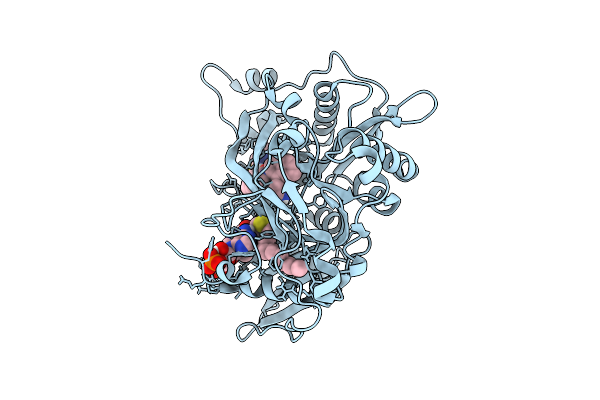
Crystal Structure Of Cryptosporidium Parvum N-Myristoyltransferase With Bound Myristoyl-Coa And Inhibitor 20057
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE/INHIBITOR Ligands: MYA, A1BHQ, CL, 1PE, PG4 |
 |
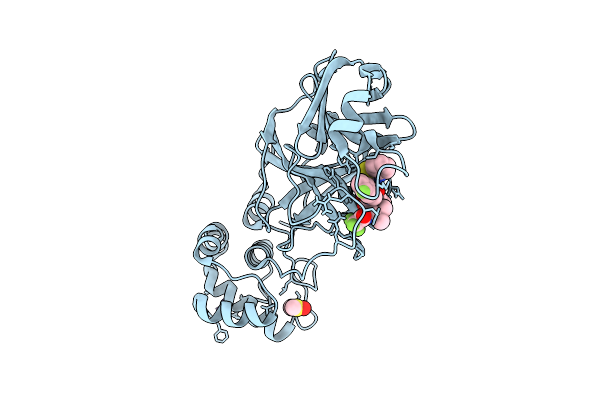
Crystal Structure Of Cryptosporidium Parvum N-Myristoyltransferase With Bound Myristoyl-Coa And Inhibitor 20084
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE/INHIBITOR Ligands: MYA, A1BHR, CL |
 |
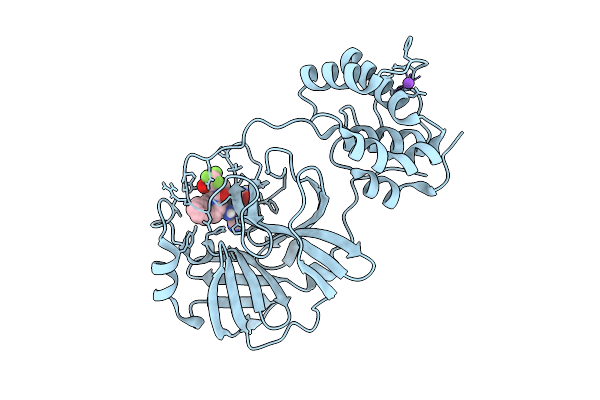
X-Ray Structure Of Sars-Cov-2 Main Protease Covalently Bound To Compound Grl-050-23 At 1.6 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN Ligands: DMS, A1BMU, NA |
 |
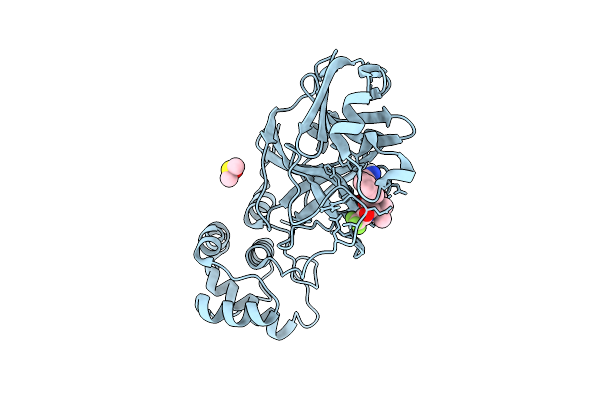
X-Ray Structure Of Sars-Cov-2 Main Protease M49I Covalently Bound To Inhibitor Grl-051-22 At 1.50 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN Ligands: A1BFE, NA |
 |
X-Ray Structure Of Sars-Cov-2 Main Protease V186F Covalently Bound To Inhibitor Grl-051-22 At 1.60 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN, HYDROLASE Ligands: A1BFE, DMS |
 |
Organism: Mus musculus, Orthomarburgvirus marburgense
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: VIRAL PROTEIN Ligands: NAG |
 |
X-Ray Structure Of Sars-Cov-2 Main Protease V186I Covalently Bound To Inhibitor Grl-051-22 At 1.90 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN, HYDROLASE Ligands: A1BFE, K |
 |
X-Ray Structure Of Sars-Cov-2 Main Protease V186F Covalently Bound To Inhibitor Grl-050-23 At 1.55 A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN, HYDROLASE Ligands: A1BMU |

