Search Count: 75
 |
Crystal Structure Of Acyl-Coa Lyase Subunit Beta From Pseudomonas Aeruginosa Pao1
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: LYASE |
 |
Crystal Structure Of A Tartrate Dehydratase From Azospirillum, Target Efi-502395, With Bound Mg And A Putative Acrylate Ion, Ordered Active Site
Organism: Azospirillum sp. b510
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2013-05-15 Classification: LYASE Ligands: MG, CL, AKR |
 |
Crystal Structure Of A 4-Hydroxyproline Epimerase From Burkholderia Multivorans, Target Efi-506479, With Bound Phosphate, Closed Domains
Organism: Burkholderia multivorans
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2013-05-01 Classification: LYASE Ligands: PO4, CL, GOL, NA |
 |
Crystal Structure Of An Enolase (Putative Galactarate Dehydratase, Target Efi-500740) From Agrobacterium Radiobacter, Bound Na And L-Malate, Ordered Active Site
Organism: Agrobacterium radiobacter
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2013-04-03 Classification: LYASE |
 |
Crystal Structure Of An Enolase (Putative Galactarate Dehydratase, Target Efi-500740) From Agrobacterium Radiobacter, Bound Sulfate, No Metal Ion, Ordered Active Site
Organism: Agrobacterium radiobacter k84
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2013-04-03 Classification: LYASE Ligands: SO4, EDO, CL |
 |
Crystal Structure Of An Enolase (Mandelate Racemase Subgroup, Target Efi-502087) From Agrobacterium Tumefaciens, With Bound Mg And D-Ribonohydroxamate, Ordered Loop
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-10-31 Classification: LYASE Ligands: CA, CL, MG, GOL, 0YR |
 |
Crystal Structure Of An Enolase Family Member From Vibrio Harveyi (Efi-Target 501692) With Homology To Mannonate Dehydratase, With Mg, Ethylene Glycol And Sulfate Bound (Ordered Loops, Space Group P41212)
Organism: Vibrio harveyi
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-08-29 Classification: LYASE Ligands: MG, SO4, CL, EDO, NA |
 |
Crystal Structure Of An Enolase Family Member From Vibrio Harveyi (Efi-Target 501692) With Homology To Mannonate Dehydratase, With Mg, Glycerol And Dicarboxylates Bound (Mixed Loops, Space Group I4122)
Organism: Vibrio harveyi
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-08-29 Classification: LYASE Ligands: CL, GOL, MLA, MG, MAE |
 |
Crystal Structure Of A Glucarate Dehydratase Related Protein, From Actinobacillus Succinogenes, Target Efi-502312, With Sodium And Sulfate Bound, Ordered Loop
Organism: Actinobacillus succinogenes
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-08-15 Classification: LYASE Ligands: SO4, NA, GOL, DTU, DTT |
 |
Crystal Structure Of D-Mannonate Dehydratase Homolog From Chromohalobacter Salexigens (Target Efi-502114), With Bound Na, Ordered Loop
Organism: Chromohalobacter salexigens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-06-27 Classification: LYASE |
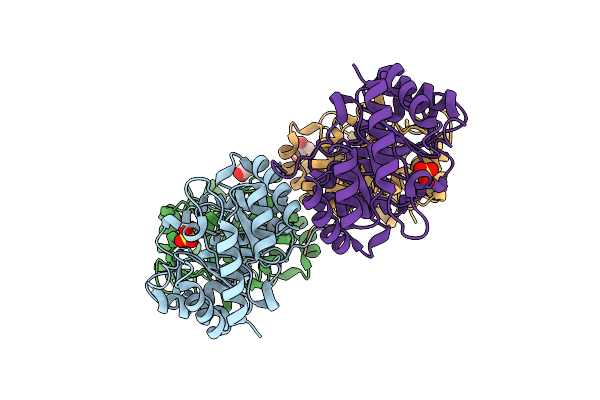 |
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2010-03-31 Classification: LYASE Ligands: SO4, GOL, TRS |
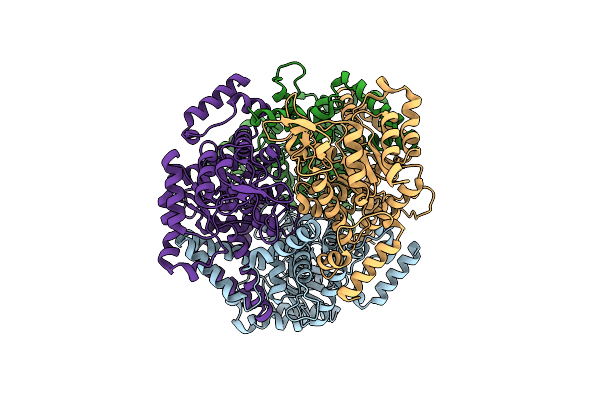 |
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Release Date: 2009-07-14 Classification: LYASE |
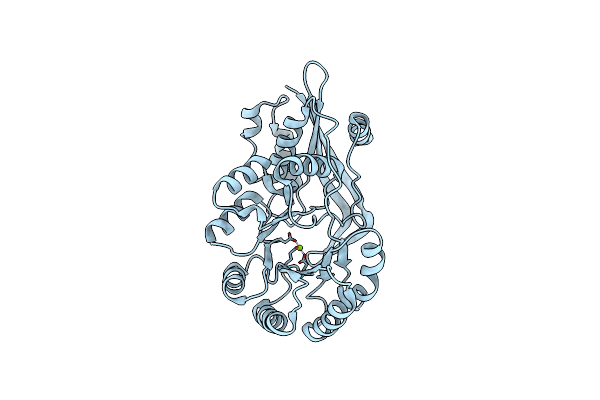 |
Crystal Structure Of O-Succinylbenzoate Synthase From Thermosynechococcus Elongatus Bp-1 Complexed With Mg In The Active Site
Organism: Thermosynechococcus elongatus bp-1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2009-05-12 Classification: LYASE Ligands: MG |
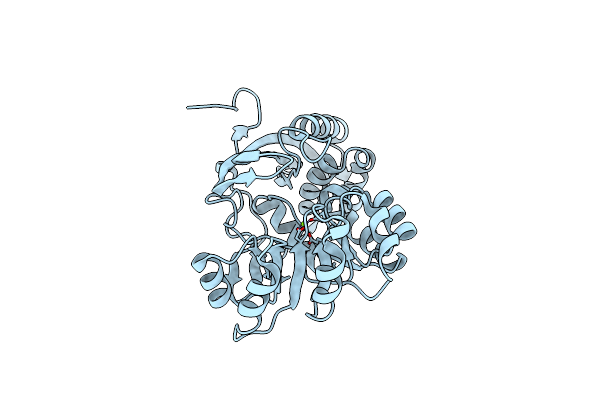 |
Crystal Structure Of O-Succinylbenzoic Acid Synthetase From Staphylococcus Aureus Complexed With Mg In The Active Site
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2009-05-05 Classification: LYASE Ligands: MG |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2008-12-09 Classification: LYASE |
 |
Crystal Structure Of Dihydrodipicolinate Synthase From Rhodopseudomonas Palustris At 2.0A Resolution
Organism: Rhodopseudomonas palustris
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2008-09-02 Classification: LYASE Ligands: PGE |
 |
Crystal Structure Of Dihydrodipicolinate Synthase From Rhodopseudomonas Palustris At 1.87A Resolution
Organism: Rhodopseudomonas palustris
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2008-08-12 Classification: LYASE |
 |
Crystal Structure Of L-Rhamnonate Dehydratase From Salmonella Typhimurium Complexed With Mg And L-Tartrate
Organism: Salmonella typhimurium lt2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-06-03 Classification: LYASE Ligands: MG, TLA |
 |
Crystal Structure Of L-Rhamnonate Dehydratase From Salmonella Typhimurium Complexed With Mg And D-Malate
Organism: Salmonella typhimurium lt2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-06-03 Classification: LYASE Ligands: MG, MLT |
 |
Crystal Structure Of Dihydrodipicolinate Synthase From Oceanobacillus Iheyensis At 1.9 A Resolution
Organism: Oceanobacillus iheyensis hte831
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-05-13 Classification: LYASE |

