Search Count: 87
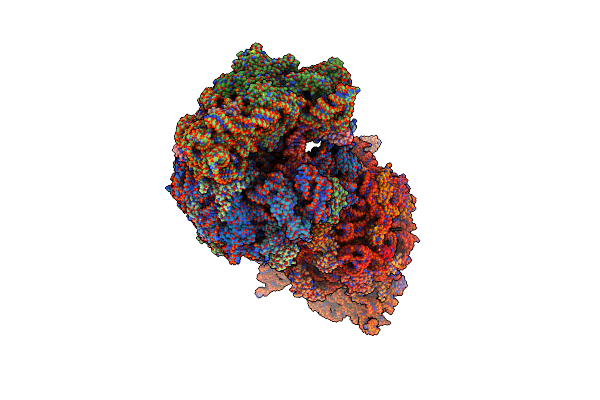 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Lasso Peptide Lariocidin, Mrna, Aminoacylated A-Site Phe-Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.50A Resolution
Organism: Escherichia coli, Paenibacillus, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG, K, ZN, SF4 |
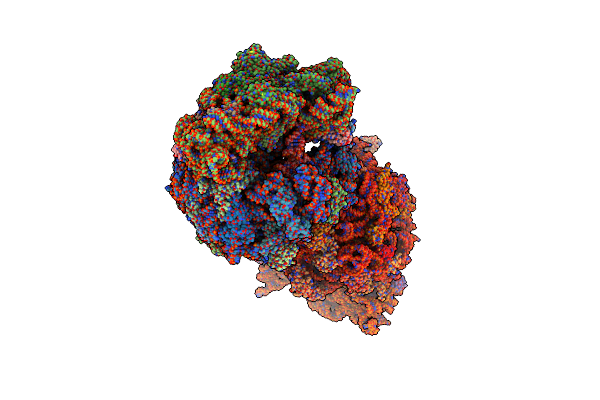 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Lasso Peptide Lariocidin B, Mrna, Aminoacylated A-Site Phe-Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.60A Resolution
Organism: Escherichia coli, Paenibacillus, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG, K, ZN, SF4 |
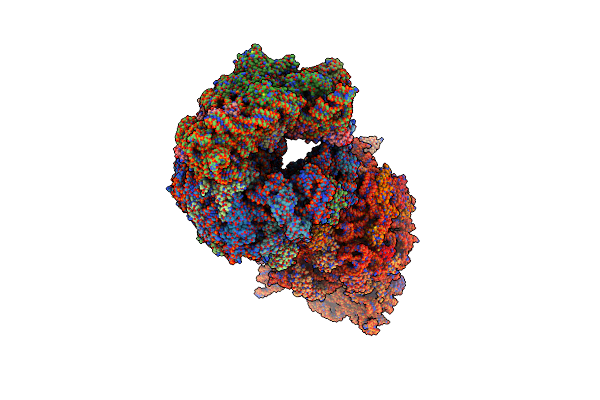 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Lasso Peptide Lariocidin And Protein Y At 2.60A Resolution
Organism: Escherichia coli k-12, Paenibacillus, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG, ARG, MPD, ZN, SF4 |
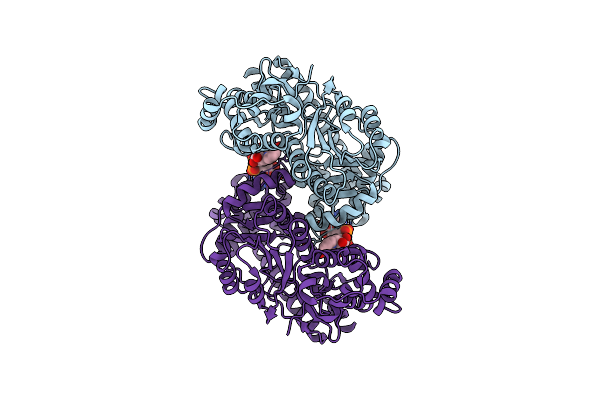 |
Organism: Pseudomonas fluorescens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: LYASE Ligands: NAD |
 |
Organism: Pseudomonas fluorescens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: LYASE |
 |
Organism: Streptomyces sp. cb03234
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-10-18 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Streptomyces sp. cb03234
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-10-18 Classification: BIOSYNTHETIC PROTEIN Ligands: 9VG |
 |
The Nmr Structure Of Noursin, A Tricyclic Ribosomal Peptide Containing A Histidine-To-Butyrine Crosslink
Organism: Streptomyces noursei atcc 11455
Method: SOLUTION NMR Release Date: 2023-05-31 Classification: UNKNOWN FUNCTION |
 |
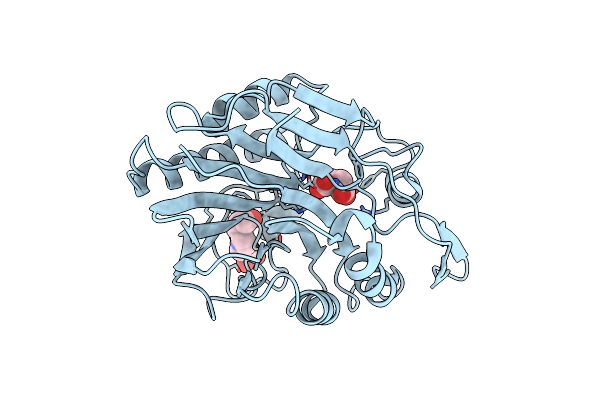
Organism: Streptomyces noursei atcc 11455
Method: SOLUTION NMR Release Date: 2023-05-31 Classification: UNKNOWN FUNCTION |
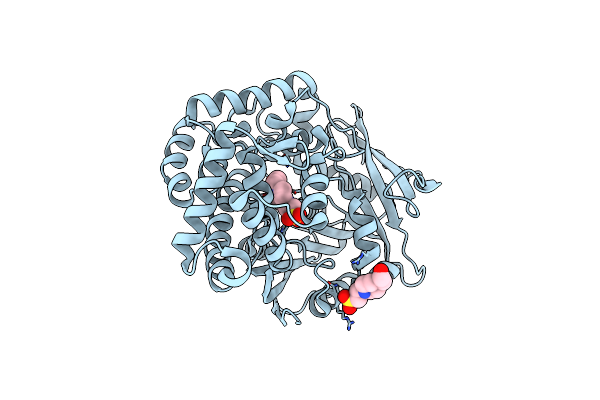 |
Organism: Streptomyces sp. cb03234
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2022-11-09 Classification: HYDROLASE Ligands: EPE |
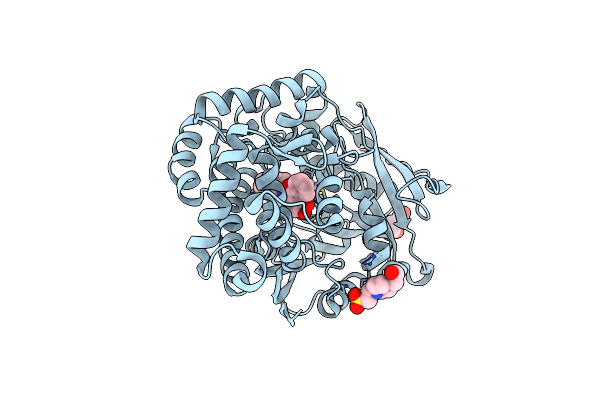 |
Organism: Streptomyces sp. cb03234
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2022-11-09 Classification: HYDROLASE Ligands: U8L, EPE, SIN |
 |
Organism: Streptomyces griseocarneus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-10-28 Classification: HYDROLASE Ligands: EPE, SIN |
 |
Organism: Hypocrea rufa
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-09-23 Classification: OXIDOREDUCTASE Ligands: FAD, LYS, GOL, EPE |
 |
 |
 |
Organism: Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2016-09-07 Classification: HYDROLASE |
 |
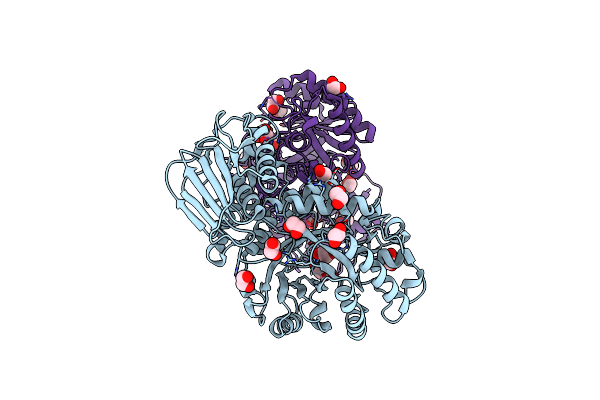 |
Structure And Activity Of The Gh20 Beta-N-Acetylhexosaminidase From Streptomyces Coelicolor A3(2)
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2014-03-12 Classification: HYDROLASE Ligands: EDO |
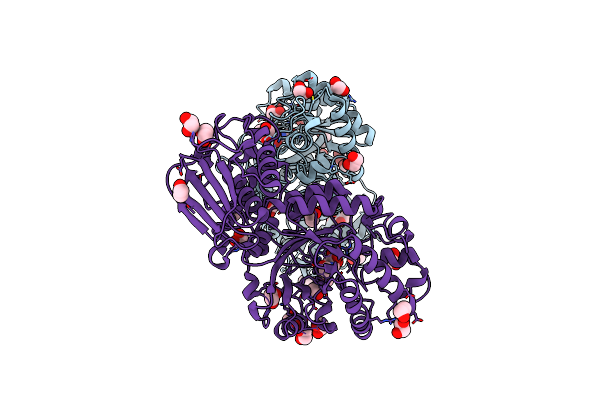 |
Structure And Activity Of The Gh20 Beta-N-Acetylhexosaminidase From Streptomyces Coelicolor A3(2)
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-03-12 Classification: HYDROLASE Ligands: EDO, GC2 |
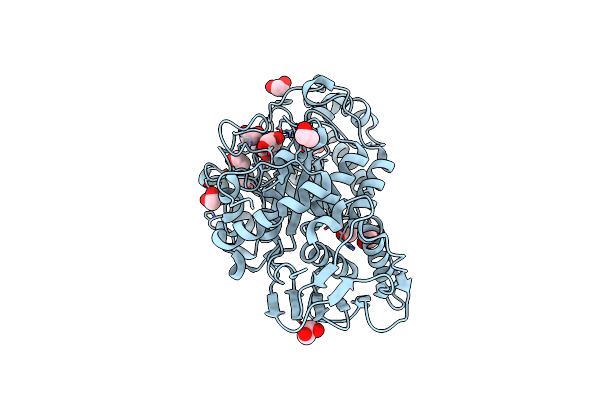 |
Structure And Activity Of The Gh20 Beta-N-Acetylhexosaminidase From Streptomyces Coelicolor A3(2)
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-03-12 Classification: HYDROLASE Ligands: NGO, EDO |

