Search Count: 25
 |
Organism: Staphylococcus aureus subsp. aureus n315
Method: SOLUTION NMR Release Date: 2025-10-15 Classification: DNA BINDING PROTEIN |
 |
Organism: Staphylococcus aureus subsp. aureus n315, Synthetic construct
Method: SOLUTION NMR Release Date: 2025-10-15 Classification: DNA BINDING PROTEIN |
 |
Organism: Vicugna pacos, Mus musculus, Escherichia coli, Staphylococcus aureus subsp. aureus n315, Streptococcus sp. 'group g'
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Staphylococcus aureus subsp. aureus n315, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-07-17 Classification: TOXIN Ligands: EPE, MPO |
 |
Organism: Staphylococcus aureus subsp. aureus n315
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-02-07 Classification: PROTEIN BINDING Ligands: ACE, GOL |
 |
Crystal Structure Of S. Aureus Gyrase In Complex With 4-[[1-[(1-Chloro-6,7-Dihydro-5H-Cyclopenta[C]Pyridin-6-Yl)Methyl]Azetidin-3-Yl]Methylamino]-6-Fluorochromen-2-One
Organism: Staphylococcus aureus subsp. aureus n315, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2023-06-28 Classification: ISOMERASE Ligands: MN, CL, ACT, 61I |
 |
Crystal Structure Of S. Aureus Gyrase In Complex With 6-[5-[2-[(4-Chloro-2,3-Dihydro-1H-Inden-2-Yl)Methylamino]Ethyl]-2-Oxo-1,3-Oxazolidin-3-Yl]-4H-Pyrido[3,2-B][1,4]Oxazin-3-One
Organism: Staphylococcus aureus subsp. aureus n315, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2023-06-28 Classification: ISOMERASE Ligands: MN, CL, 6I0 |
 |
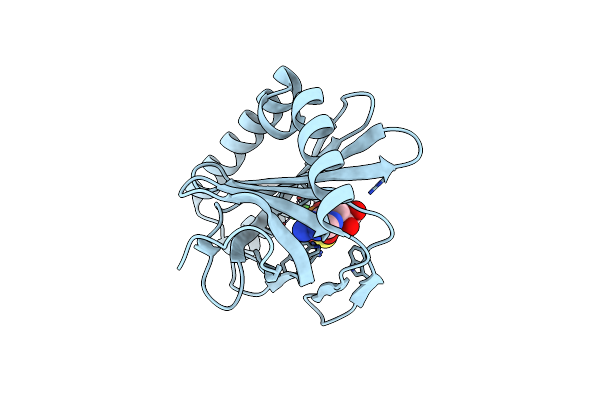
The Crystal Structure Of Nucleotide Phosphatase Sa1684 From Staphylococcus Aureus
Organism: Staphylococcus aureus subsp. aureus n315
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-03-17 Classification: CYTOSOLIC PROTEIN Ligands: CIT, GOL, MG |
 |
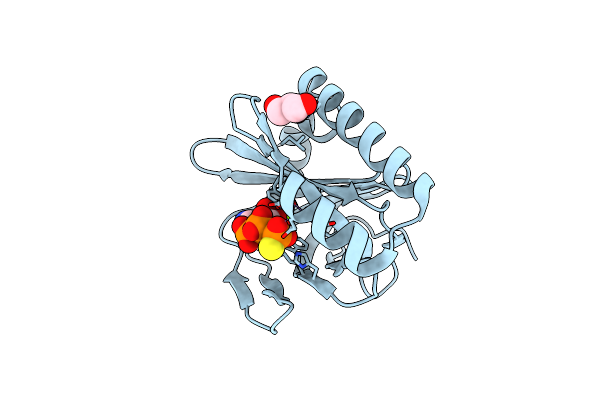
Crystal Structure Of Nucleoside Phosphatase Sa1684 Complex With Atp Analogue From Staphylococus Aureus
Organism: Staphylococcus aureus subsp. aureus n315
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2021-03-17 Classification: CYTOSOLIC PROTEIN Ligands: AGS, CA |
 |
The Structure Of Nucleoside Phosphatase Sa1684 Complex With Gtp Analogue From Staphylococcus Aureus
Organism: Staphylococcus aureus subsp. aureus n315
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2021-03-17 Classification: CYTOSOLIC PROTEIN Ligands: GSP, MG, GOL |
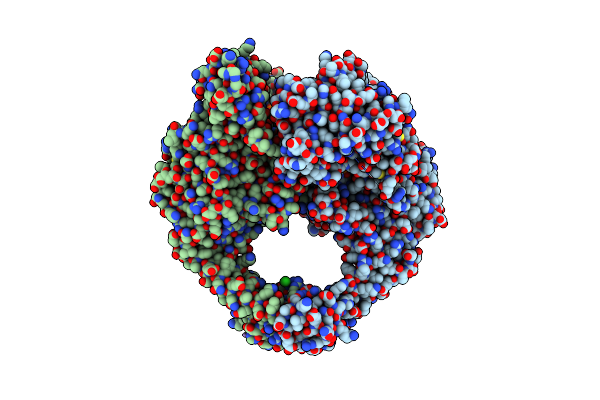 |
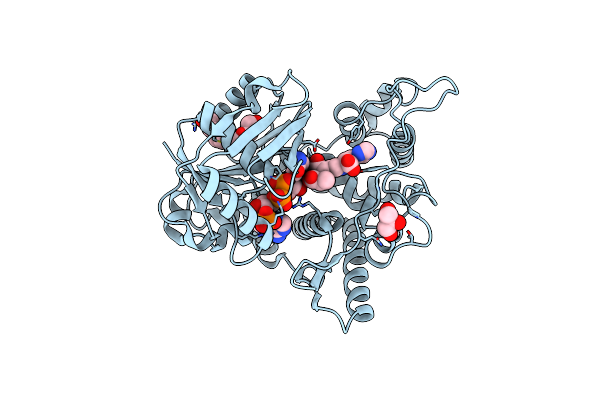
Organism: Staphylococcus aureus, Staphylococcus aureus subsp. aureus n315
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-11-11 Classification: ISOMERASE Ligands: EPE, MN, DMS, CL, Q52 |
 |
Organism: Staphylococcus aureus subsp. aureus n315
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2019-04-10 Classification: BIOSYNTHETIC PROTEIN Ligands: NAP, 8UX, GOL, PG4 |
 |
Organism: Staphylococcus aureus subsp. aureus n315
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-10-24 Classification: TRANSFERASE Ligands: FQ8, MG, CL |
 |
Organism: Staphylococcus aureus subsp. aureus n315
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2018-09-26 Classification: TRANSFERASE Ligands: CL, MG |
 |
Organism: Staphylococcus aureus subsp. aureus n315
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-09-26 Classification: TRANSFERASE Ligands: CL, UD1, MN, MG, GOL |
 |
Organism: Staphylococcus aureus subsp. aureus n315, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2018-04-04 Classification: ISOMERASE Ligands: MN, SO4, E32, GOL |
 |
Crystal Structure Of S. Aureus Fabi In Complex With Nadp And 4-Fluoro- 5-Hexyl-2-Phenoxyphenol
Organism: Staphylococcus aureus subsp. aureus n315
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2015-03-04 Classification: OXIDOREDUCTASE Ligands: GLU, W0I, NAP |
 |
Crystal Structure Of S. Aureus Fabi In Complex With Nadp And 2-(2- Chloro-4-Nitrophenoxy)-5-Ethyl-4-Fluorophenol
Organism: Staphylococcus aureus subsp. aureus n315
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2015-03-04 Classification: OXIDOREDUCTASE Ligands: GLU, MRD, NAP, 9W7 |
 |
Crystal Structure Of S. Aureus Fabi In Complex With Nadp And 5-Ethyl- 4-Fluoro-2-((2-Fluoropyridin-3-Yl)Oxy)Phenol
Organism: Staphylococcus aureus subsp. aureus n315
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-03-04 Classification: OXIDOREDUCTASE Ligands: NAP, MRD, JA3, GLU |
 |
Crystal Structure Of S. Aureus Fabi In Complex With Nadp And 5-Bromo- 2-(4-Chloro-2-Hydroxyphenoxy)Benzonitrile
Organism: Staphylococcus aureus subsp. aureus n315
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2015-03-04 Classification: OXIDOREDUCTASE Ligands: GLU, NAP, J47 |

