Search Count: 289
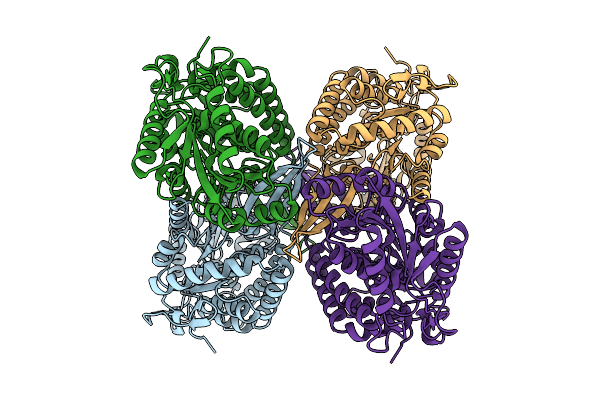 |
Cryoem Structure Of Aldehyde Dehydrogenase From Burkholderia Cenocepacia At 2.33A Resolution
Organism: Burkholderia cenocepacia
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: OXIDOREDUCTASE |
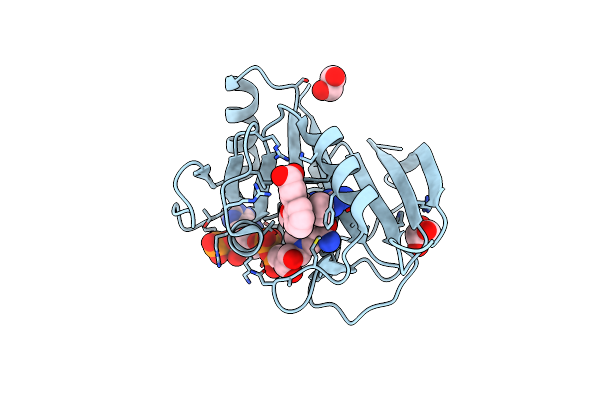 |
Crystal Structure Of A Dihydrofolate Reductase Fola From Stenotrophomonas Maltophilia Bound To Nadp And P218
Organism: Stenotrophomonas maltophilia k279a
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-11-23 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: NAP, MMV, EDO, GOL |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2021-10-06 Classification: TRANSFERASE Ligands: PO4, CL, MG |
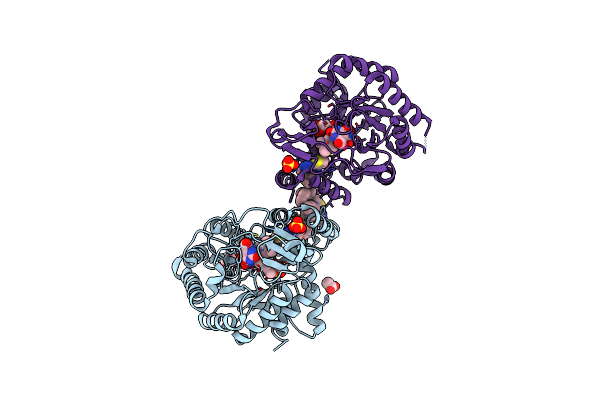 |
Crystal Structure Of Dihydroorotate Dehydrogenase From Plasmodium Falciparum In Complex With Orotate, Fmn, And Inhibitor Ncgc00600348-01
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2021-09-29 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: 8IE, FMN, ORO, ACT, P4K, SO4 |
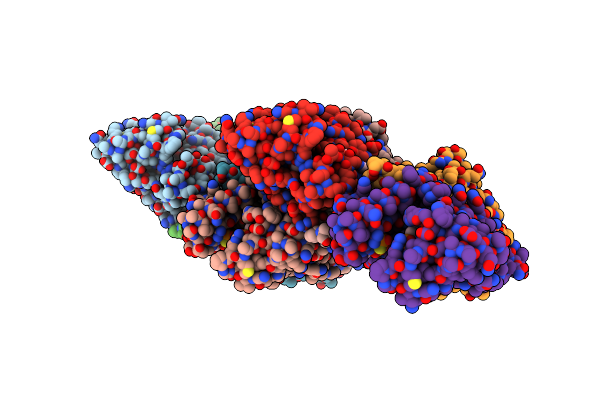 |
Crystal Structure Of A Guanylate Kinase From Stenotrophomonas Maltophilia K279A With Heterogeneous Ligand States Of Gmp/Adp, Gmp/-, Gdp/-, And Gmp/Atpgs
Organism: Stenotrophomonas maltophilia (strain k279a)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-09-22 Classification: TRANSFERASE Ligands: 5GP, ADP, GDP, AGS |
 |
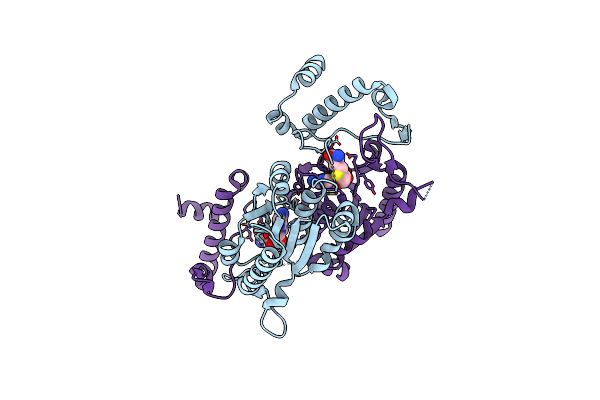
Crystal Structure Of A Trna (Guanine-N1)-Methyltransferase From Acinetobacter Baumannii
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2021-06-09 Classification: TRANSFERASE |
 |
Crystal Structure Of A Trna (Guanine-N1)-Methyltransferase From Acinetobacter Baumannii Ab5075-Uw Bound To S-Adenosyl Homocysteine
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2021-06-09 Classification: TRANSFERASE Ligands: SAH |
 |
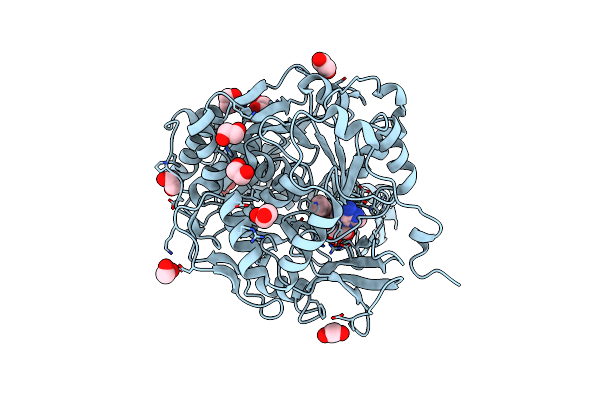
Organism: Legionella pneumophila subsp. pneumophila (strain philadelphia 1 / atcc 33152 / dsm 7513)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-10-28 Classification: HYDROLASE Ligands: EDO, NAD, SIN |
 |
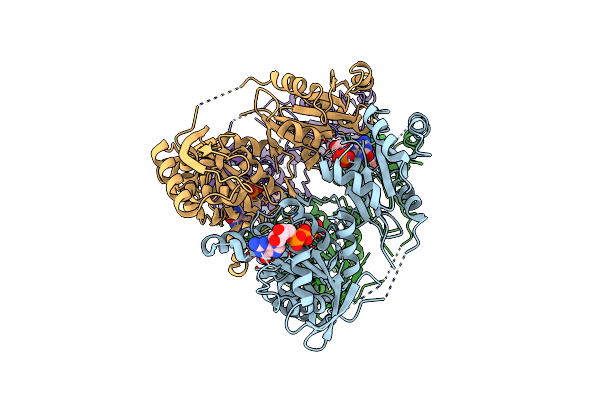
Crystal Structure Of Acetyl-Coa Synthetase In Complex With Adenosine-5'-Propylphosphate From Coccidioides Posadasii C735
Organism: Coccidioides posadasii (strain c735)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2020-10-21 Classification: LIGASE Ligands: PRX, MG, EDO |
 |
Crystal Structure Of A Gtp-Binding Protein Enga (Der Homolog) From Neisseria Gonorrhoeae Bound To Gdp
Organism: Neisseria gonorrhoeae (strain nccp11945)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-09-23 Classification: HYDROLASE Ligands: GDP, IOD, CL |
 |
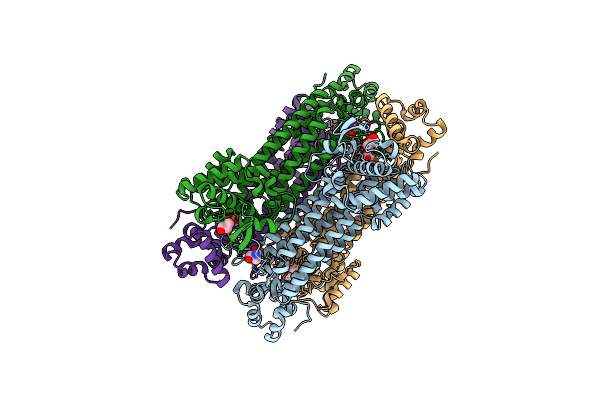
Crystal Structure Of Influenza A Virus Hemagglutinin From A/Ohio/09/2015 Bound To The Stalk-Binding Cr6261 Antibody Fab
Organism: Influenza a virus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2020-09-02 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, CL |
 |
Crystal Structure Of An Aspartate Ammonia-Lyase From Elizabethkingia Anophelis Nuhp1
Organism: Elizabethkingia anophelis nuh1
Method: X-RAY DIFFRACTION Release Date: 2020-04-29 Classification: LYASE Ligands: TRS, FUM, GOL, CL |
 |
Crystal Structure Of Uroporphyrinogen Iii Decarboxylate (Heme) From Stenotrophomonas Maltophilia
Organism: Stenotrophomonas maltophilia (strain k279a)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-04-22 Classification: LYASE Ligands: EDO, SO4, CL |
 |
Crystal Structure Of A Pyridoxine 5'-Phosphate Synthease From Stenotrophomonas Maltophilia K279A
Organism: Stenotrophomonas maltophilia (strain k279a)
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2020-03-25 Classification: LYASE Ligands: PO4 |
 |
Crystal Structure Of A Ribosome Recycling Factor From Ehrlichia Chaffeensis
Organism: Ehrlichia chaffeensis (strain atcc crl-10679 / arkansas)
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2020-03-18 Classification: TRANSLATION Ligands: CL |
 |
Crystal Structure Of Threonyl-Trna Synthetase (Thrrs) From Stenotrophomonas Maltophilia K279A
Organism: Stenotrophomonas maltophilia (strain k279a)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-03-04 Classification: LIGASE Ligands: SO4, ZN |
 |
Crystal Structure Of A Dnan Sliding Clamp (Dna Polymerase Iii Subunit Beta) From Pseudomonas Aeruginosa Bound To Griselimycin
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1), Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2019-08-07 Classification: TRANSFERASE/ANTIBIOTIC Ligands: SO4 |
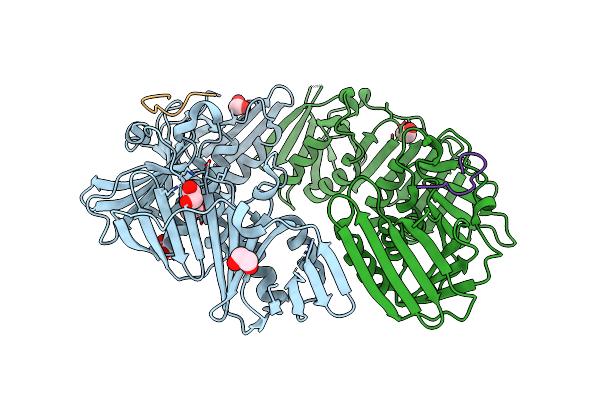 |
Crystal Structure Of A Dnan Sliding Clamp (Dna Polymerase Iii Subunit Beta) From Bartonella Birtlesii Bound To Griselimycin
Organism: Bartonella birtlesii ll-wm9, Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-08-07 Classification: TRANSFERASE/ANTIBIOTIC Ligands: EDO, IOD |
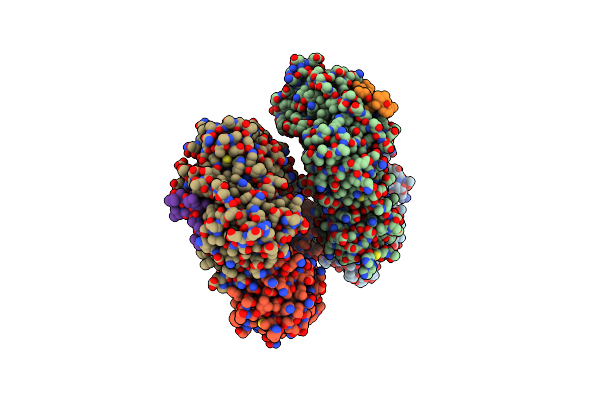 |
Crystal Structure Of A Dnan Sliding Clamp (Dna Polymerase Iii Subunit Beta) From Rickettsia Rickettsii Bound To Griselimycin
Organism: Rickettsia rickettsii (strain sheila smith), Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-08-07 Classification: TRANSFERASE/ANTIBIOTIC Ligands: MPD |
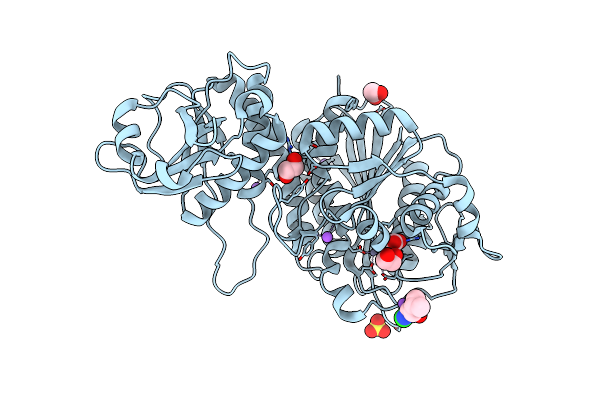 |
Crystal Structure Of A Probable Cytosol Aminopeptidase (Leucine Aminopeptidase, Lap) From Chlamydia Trachomatis D/Uw-3/Cx
Organism: Chlamydia trachomatis (strain d/uw-3/cx)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-05-01 Classification: HYDROLASE Ligands: CO3, EDO, SO4, ZN, NA, CL, LEU |

