Search Count: 22
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2023-07-26 Classification: TRANSFERASE Ligands: HQG, GOL, PO4, EPE, GLY, SOR |
 |
Crystal Structure Of Glucose-2-Epimerase In Complex With D-Glucitol From Runella Slithyformis Runsl_4512
Organism: Runella slithyformis
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2023-07-12 Classification: ISOMERASE Ligands: SOR |
 |
Crystal Structure Of Glucose-2-Epimerase Mutant_D254A In Complex With D-Glucitol From Runella Slithyformis Runsl_4512
Organism: Runella slithyformis
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2023-07-12 Classification: ISOMERASE Ligands: SOR, FMT |
 |
Structure Of The Oxomolybdenum Mesoporphyrin Ix-Reconstituted Cyp102A1 Haem Domain With N-Enanthyl-L-Prolyl-L-Phenylalanine In Complex With Styerene
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: MI9, D0L, SYN, SOR |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-05-26 Classification: CELL ADHESION Ligands: SOR |
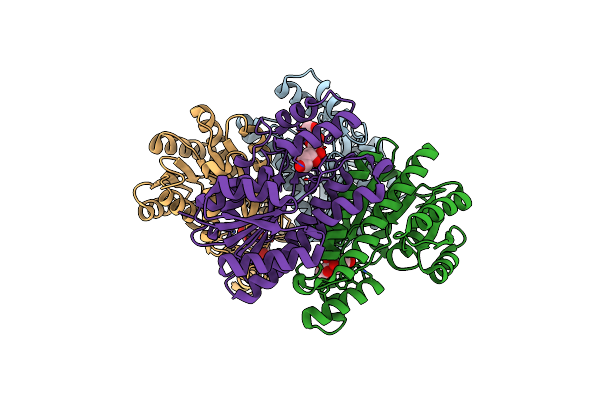 |
Structure Of Sorbitol Dehydrogenase From Sinorhizobium Meliloti 1021 Bound To Sorbitol
Organism: Sinorhizobium meliloti 1021
Method: X-RAY DIFFRACTION Release Date: 2020-06-24 Classification: OXIDOREDUCTASE Ligands: SOR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-02-07 Classification: HYDROLASE Ligands: CA, CL, E64, SOR |
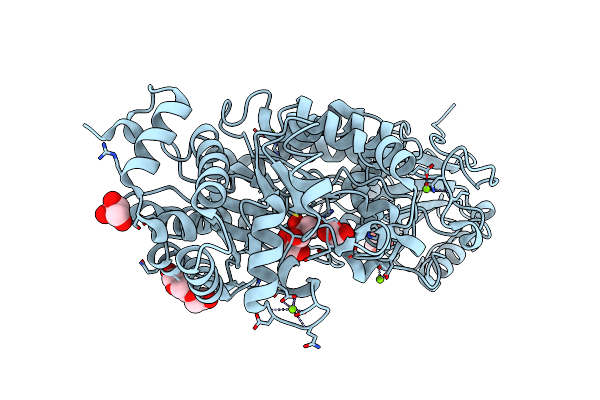 |
Crystal Structure Glucan 1,4-Beta-Glucosidase From Saccharopolyspora Erythraea
Organism: Saccharopolyspora erythraea d
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2017-11-29 Classification: HYDROLASE Ligands: MG, GOL, PGE, SOR |
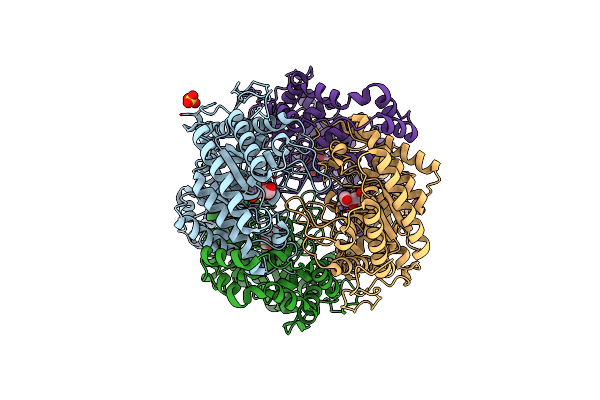 |
Crystal Structure Of Xylose Isomerase From Piromyces Sp. E2 In Complex With Two Mn2+ Ions And Sorbitol
Organism: Piromyces sp. e2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-11-01 Classification: ISOMERASE Ligands: MN, SOR, SO4 |
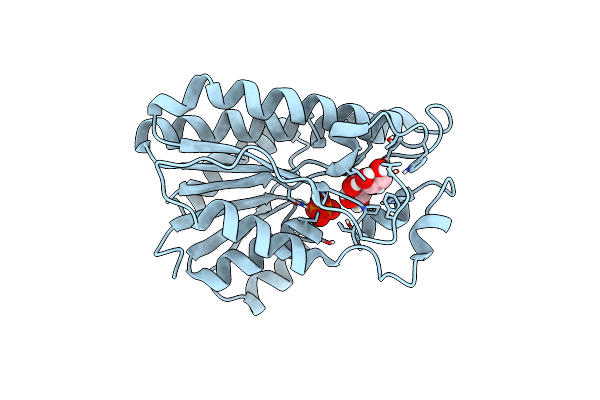 |
Structural Characterization Of The Thermostable Bradyrhizobium Japonicum D-Sorbitol Dehydrogenase
Organism: Bradyrhizobium japonicum
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2016-11-09 Classification: OXIDOREDUCTASE Ligands: PO4, SOR |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2016-08-31 Classification: TRANSFERASE Ligands: ZN, SOR |
 |
Crystal Structure Of Aldose-Aldose Oxidoreductase From Caulobacter Crescentus Complexed With Sorbitol
Organism: Caulobacter crescentus cb15
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2015-10-21 Classification: OXIDOREDUCTASE Ligands: NDP, SOR, SO4 |
 |
Room-Temperature Joint X-Ray/Neutron Structure Of D-Xylose Isomerase In Complex With 2Ni2+ And Per-Deuterated D-Sorbitol At Ph 5.9
Organism: Streptomyces rubiginosus
Method: NEUTRON DIFFRACTION, X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-08-29 Classification: ISOMERASE Ligands: NI, SOR, DOD |
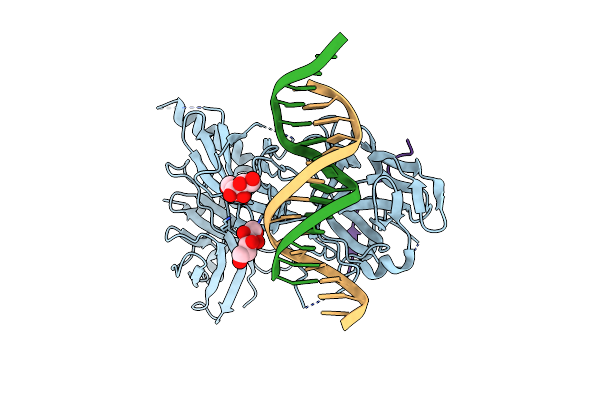 |
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2008-04-01 Classification: DNA BINDING PROTEIN/DNA Ligands: SOR |
 |
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2008-01-08 Classification: OXIDOREDUCTASE Ligands: FAD, SOR, CL |
 |
Crystal Structure Of The Complex Formed Between C-Terminal Half Of Bovine Lactoferrin And Sorbitol At 2.85 A Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2006-09-12 Classification: METAL BINDING PROTEIN Ligands: SOR, FE, CO3, ZN, SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1999-11-10 Classification: LYASE Ligands: SOR, MG, SO4, GLV |
 |
Modes Of Binding Substrates And Their Analogues To The Enzyme D-Xylose Isomerase
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1994-06-22 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: SOR, MN |
 |
Protein Engineering Of Xylose (Glucose) Isomerase From Actinoplanes Missouriensis. 1. Crystallography And Site-Directed Mutagenesis Of Metal Binding Sites
Organism: Actinoplanes missouriensis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1993-07-15 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: SOR, CO |
 |
Protein Engineering Of Xylose (Glucose) Isomerase From Actinoplanes Missouriensis. 1. Crystallography And Site-Directed Mutagenesis Of Metal Binding Sites
Organism: Actinoplanes missouriensis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 1993-07-15 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: SOR, MG |

