Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Aspergillus fumigatus
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: A1EKF, SIN, CO |
 |
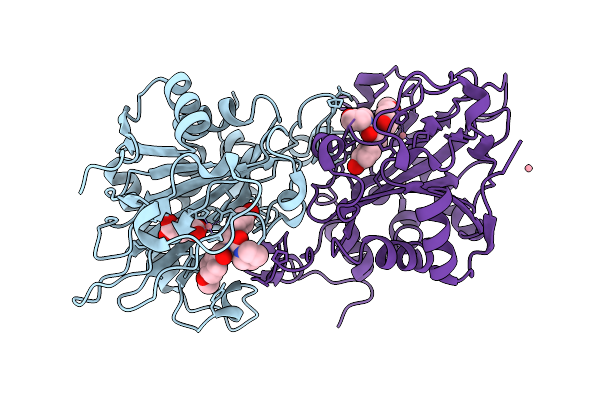
Succinyl-Coa:(R)-Benzylsuccinate Coa-Transferase (Bbsef), D178-Coa Adduct + Succinate (Weakly Occupied)
Organism: Aromatoleum aromaticum
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: TRANSFERASE Ligands: GOL, PGE, COA, TRS, SIN |
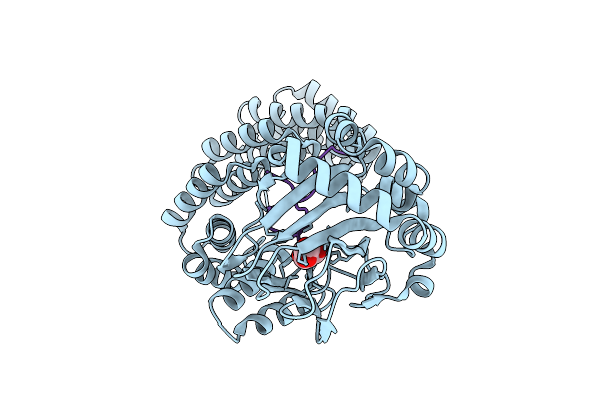 |
Aspartyl/Asparaginyl Beta-Hydroxylase (Asph) In Complex With Fe, 2Og, Sin And Hydroxylated Product Of Factor X Peptide Fragment (39Mer-4Ser)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: OXIDOREDUCTASE Ligands: SIN, AKG, FE |
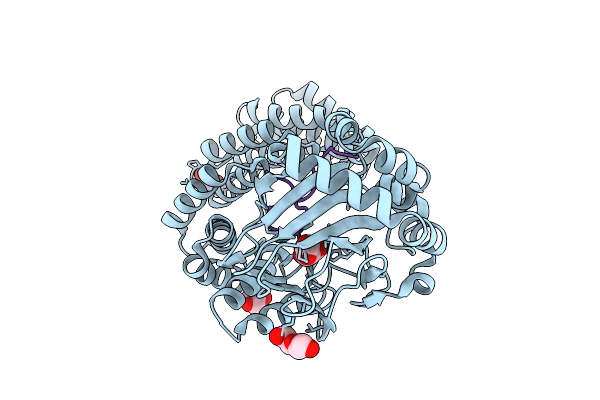 |
Aspartyl/Asparaginyl Beta-Hydroxylase (Asph) In Complex With Fe, 2-Oxoglutarate, Succinate And The Hudroxylated Product Of Factor X Derived Peptide Fragment After O2 Exposure
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: OXIDOREDUCTASE Ligands: AKG, SIN, PEG, FE, GOL, CL |
 |
Room Temperature Structure Of Aspartyl/Asparaginyl Beta-Hydroxylase (Asph) In Complex With Fe, 2-Oxoglutarate And Hydroxylated Factor X Derived Peptide Fragment, 2 H O2 Exposure
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: OXIDOREDUCTASE Ligands: SIN, FE |
 |
Aspartyl/Asparaginyl Beta-Hydroxylase (Asph) In Complex With Fe, 2-Oxoglutarate, Succinate And The Hydroxylated Product Of Factor X Derived Peptide Fragment, 12 H O2 Exposure
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: OXIDOREDUCTASE Ligands: SIN, FE |
 |
Room Temperature Structure Of Aspartyl/Asparaginyl Beta-Hydroxylase (Asph) In Complex With Fe, 2-Oxoglutarate, And Hydroxylated Factor X Derived Peptide Fragment, 1.5 S O2 Exposure
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: OXIDOREDUCTASE Ligands: AKG, SIN, FE |
 |
Crystal Structure Of Formyl-Coenzyme A Transferase From Brucella Melitensis In Complex With Succinate
Organism: Brucella melitensis 16m1w
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: TRANSFERASE Ligands: SIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: METAL BINDING PROTEIN Ligands: CA, 117, NA, MLI, ACY, SIN, FMT |
 |
Organism: Actinoplanes teichomyceticus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: BIOSYNTHETIC PROTEIN Ligands: LYS, AKG, FE2, CL, SIN, YL0, A1CA1 |
 |
Organism: Actinoplanes teichomyceticus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: BIOSYNTHETIC PROTEIN Ligands: LYS, SIN, PGE, VVO, CL, FLC, V |
 |
Organism: Streptomyces roseifaciens
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: BIOSYNTHETIC PROTEIN Ligands: VVO, SIN |
 |
Organism: Actinoplanes teichomyceticus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: BIOSYNTHETIC PROTEIN Ligands: LYS, SIN, VVO, CL |
 |
Organism: Actinoplanes teichomyceticus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: BIOSYNTHETIC PROTEIN Ligands: LYS, SIN, VVO, CL, V |
 |
Organism: Actinoplanes teichomyceticus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: BIOSYNTHETIC PROTEIN Ligands: LYS, SIN, VVO, CL, V |
 |
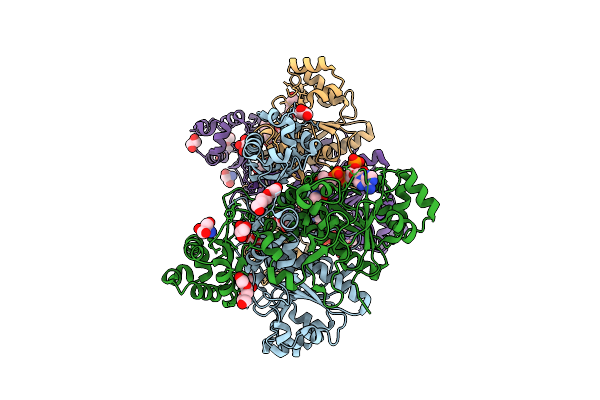
Cryo-Em Structure Of The Apo-Form Succinate Dehydrogenase From Chloroflexus Aurantiacus
Organism: Chloroflexus aurantiacus j-10-fl
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: FAD, SIN, SF4, FES, F3S, HEM, LMT, PGV, PEV |
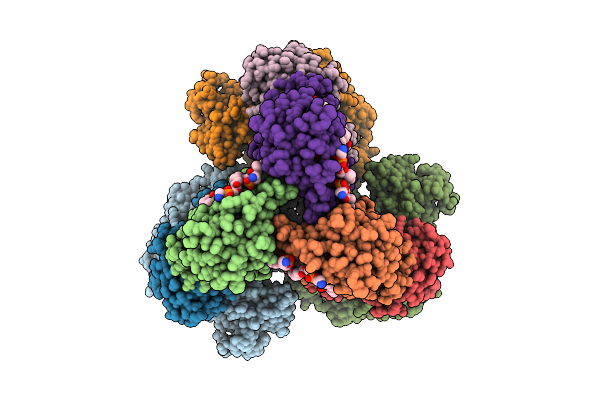 |
Cryo-Em Structure Of The Lipid-Bound Succiante Dehydrogenase From Chloroflexus Aurantiacus
Organism: Chloroflexus aurantiacus j-10-fl
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: FAD, SIN, SF4, FES, F3S, HEM, LMT, PGV, PEV |
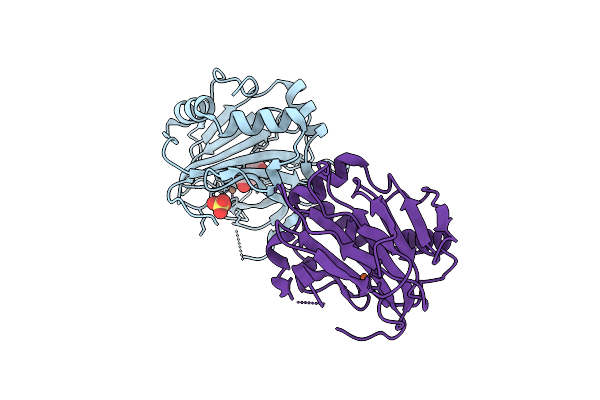 |
X-Ray Crystal Structure Of Streptomyces Cacaoi Pold With Iron And Succinate Bound
Organism: Streptomyces cacaoi
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: FE, SO4, SIN |
 |
K115 Acetylated Human Muscle Pyruvate Kinase, Isoform M2 (Pkm2), In Complex With Fbp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSFERASE Ligands: FBP, EDO, SIN, GOL, K |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSFERASE Ligands: GOL, EDO, MG, TRS, SIN, K |