Search Count: 8
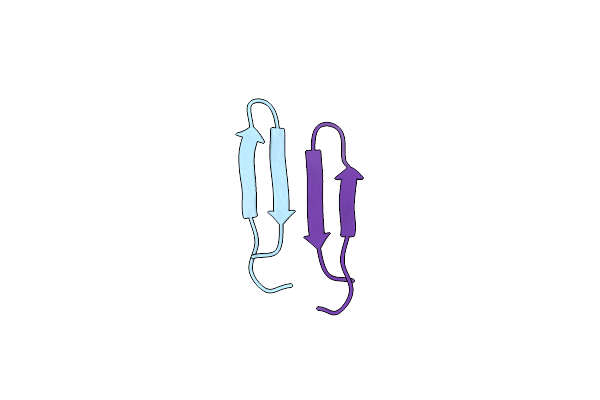 |
Membrane-Bound Dimer Structure Of Protegrin-1 (Pg-1), A Beta-Hairpin Antimicrobial Peptide In Lipid Bilayers From Rotational-Echo Double-Resonance Solid-State Nmr
|
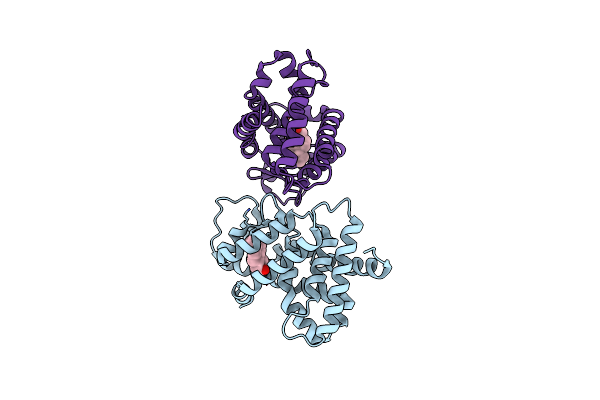 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2005-01-04 Classification: LIGAND RECEPTOR/TRANSCRIPTION REGULATION Ligands: ATE |
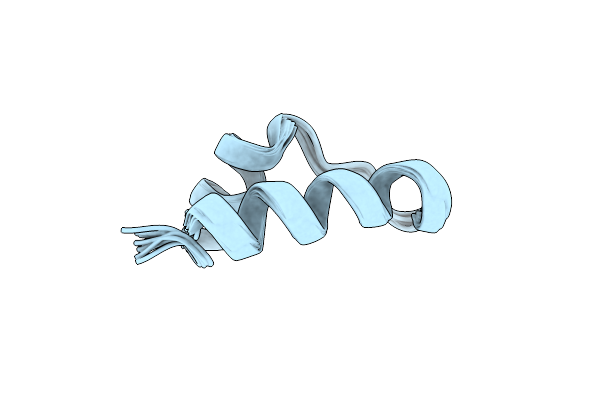 |
Conformational Mapping Of Mini-B: An N-Terminal/C-Terminal Construct Of Surfactant Protein B Using 13C-Enhanced Fourier Transform Infrared (Ftir) Spectroscopy
Organism: Homo sapiens
Method: INFRARED SPECTROSCOPY Release Date: 2004-06-15 Classification: SURFACE ACTIVE PROTEIN |
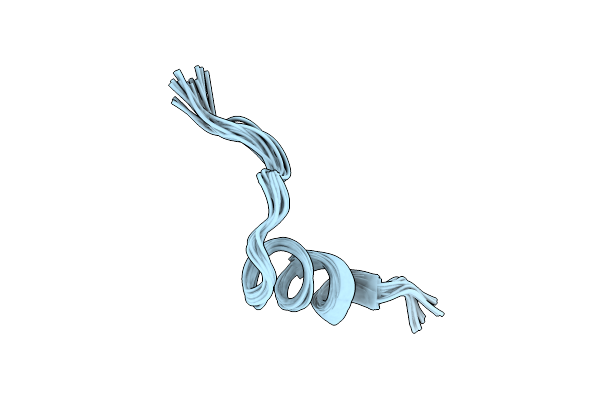 |
Conformational Mapping Of The N-Terminal Peptide Of Hiv-1 Gp41 In Lipid Detergent And Aqueous Environments Using 13C-Enhanced Fourier Transform Infrared Spectroscopy
|
 |
The Solution Structure Of Sheep Myeloid Antimicrobial Peptide, Residues 1-29 (Smap29)
|
 |
Conformational Mapping Of The N-Terminal Fusion Peptide Of Hiv-1 Gp41 Using 13C-Enhanced Fourier Transform Infrared Spectroscopy (Ftir)
|
 |
Conformational Mapping Of The N-Terminal Segment Of Surfactant Protein B In Lipid Using 13C-Enhanced Fourier Transform Infrared Spectroscopy (Ftir)
|
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1996-06-10 Classification: PHOSPHOTRANSFERASE (CARBOXYL ACCEPTOR) Ligands: MAP, 3PG |

