Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: MEMBRANE PROTEIN Ligands: SF4, FMN, PEE, 8Q1, NDP, FES, PLX, ZN, CDL, DGT, MG |
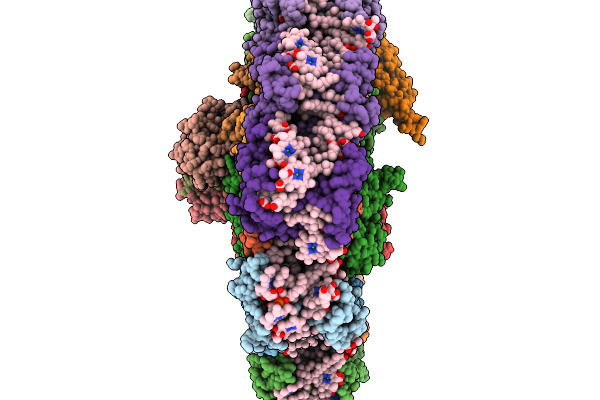 |
Organism: Chroomonas placoidea
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: PHOTOSYNTHESIS Ligands: CLA, KC2, II0, II3, IHT, LHG, LMG, SQD, 8CT, DGD, SF4, PQN |
 |
The Psi3-Isia43 Complex With A Closed Double Ring Of Isia Proteins Bound To A Trimeric Psi Core
Organism: Thermosynechococcus vestitus bp-1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: PHOTOSYNTHESIS Ligands: CLA, PQN, SF4, BCR, LHG, LMU, SQD, LMG, CA |
 |
The Psi1-Isia13 Complex With Double-Layered Isia Proteins Bound To The Monomeric Psi Core
Organism: Thermosynechococcus vestitus bp-1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: PHOTOSYNTHESIS Ligands: CLA, BCR, SQD, LMG, LHG, PQN, SF4, LMU |
 |
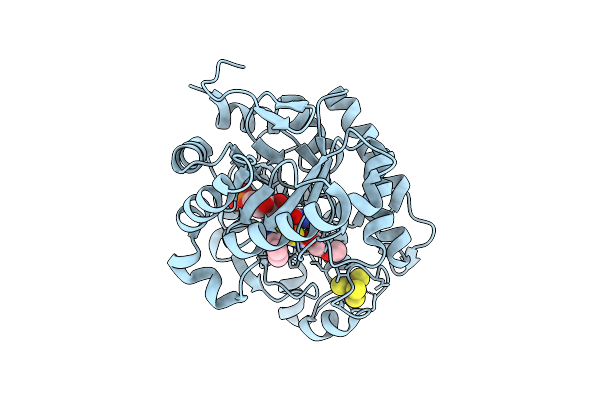
[Fefe]-Hydrogenase From D. Desulfuricans With Synthetic Active Site Containing Only One Cyanide Ligand.
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: OXIDOREDUCTASE Ligands: SF4, A1IW2, CL, LI |
 |
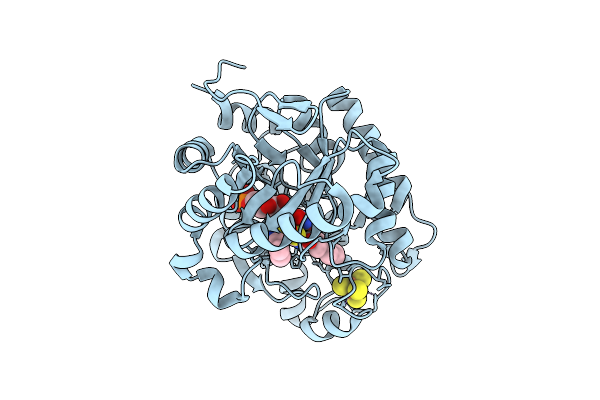
Crystal Structure Of Dehydrogenase/Isomerase Fabx From Helicobacter Pylori In Complex With Inhibitor 1
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: BIOSYNTHETIC PROTEIN Ligands: SF4, A1ELE, FMN |
 |
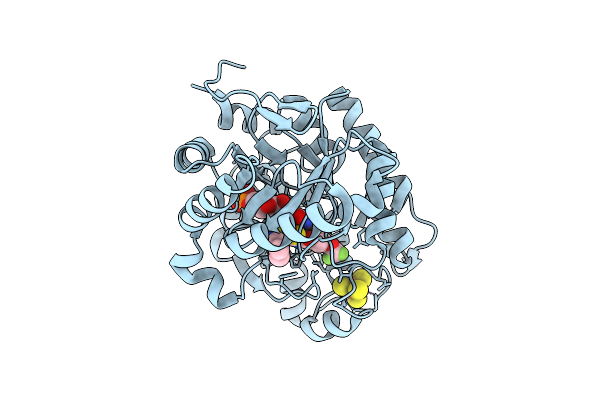
Crystal Structure Of Dehydrogenase/Isomerase Fabx From Helicobacter Pylori In Complex With Inhibitor 21
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: BIOSYNTHETIC PROTEIN Ligands: FMN, SF4, A1ELF |
 |
Crystal Structure Of Dehydrogenase/Isomerase Fabx From Helicobacter Pylori In Complex With Inhibitor 11
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: BIOSYNTHETIC PROTEIN Ligands: FMN, SF4, A1ELG |
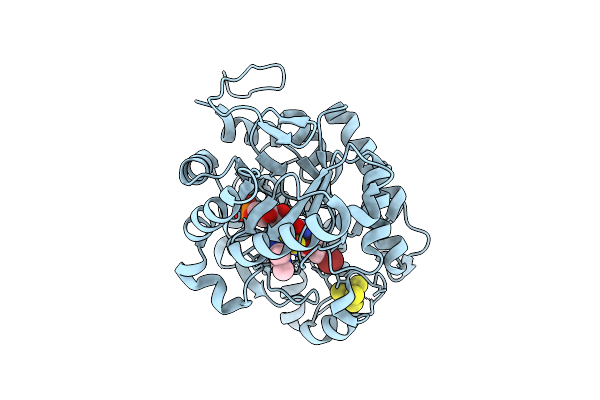 |
Crystal Structure Of Dehydrogenase/Isomerase Fabx From Helicobacter Pylori In Complex With Inhibitor 35
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: BIOSYNTHETIC PROTEIN Ligands: FMN, SF4, A1ELH |
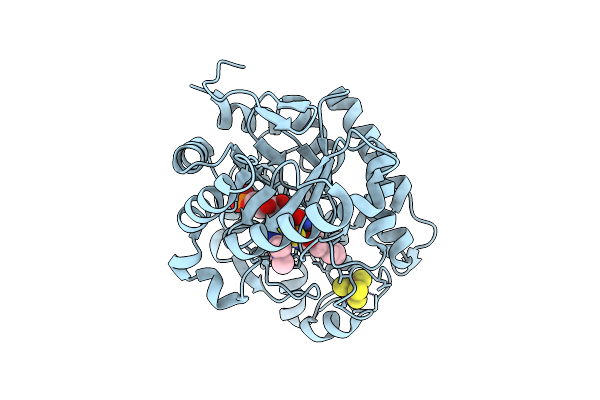 |
Crystal Structure Of Dehydrogenase/Isomerase Fabx From Helicobacter Pylori In Complex With Inhibitor 47
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: BIOSYNTHETIC PROTEIN Ligands: FMN, SF4, A1EMD |
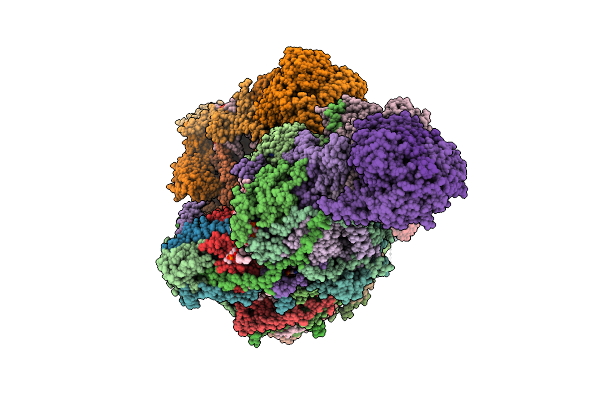 |
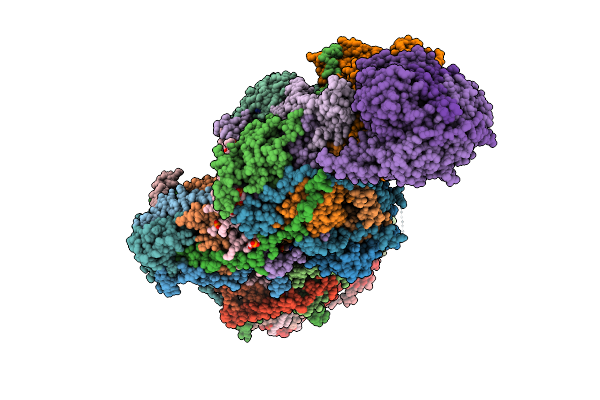 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: CDL, PEE, NDP, ZMP, ADP, 3PE, PC1, PLX, XEW, SF4, FES, MG, ZN, FMN |
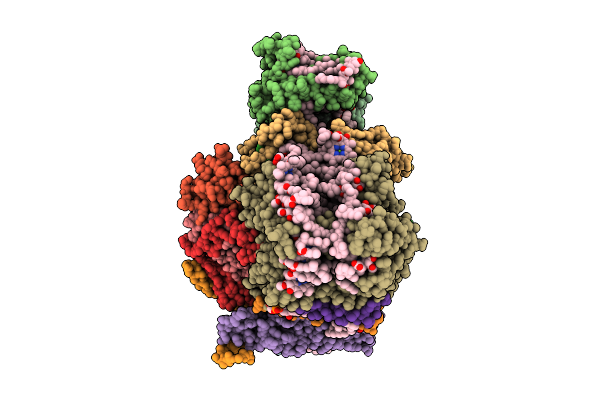 |
Structure Of The Wild-Type Psi-5Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, DGD, LMG, A1L1F, PQN, LHG, BCR, SF4 |
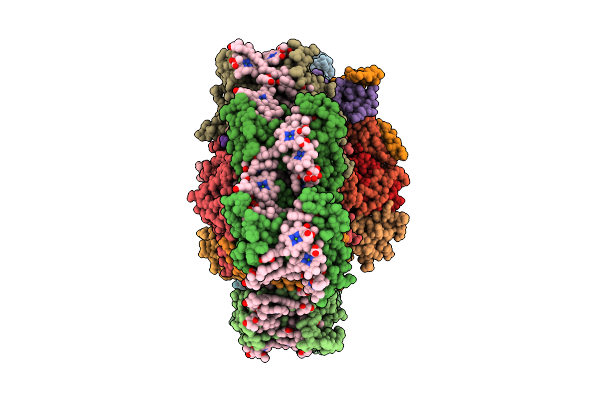 |
Structure Of The Canthaxanthin Mutant Psi-9Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, A1L1F, 45D, LHG, DGD, LMG, PQN, BCR, SF4 |
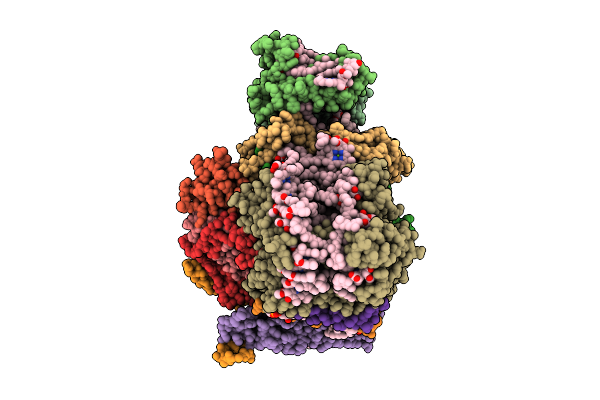 |
Structure Of The Canthaxanthin Mutant Psi-5Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, DGD, LMG, A1L1F, PQN, LHG, BCR, SF4 |
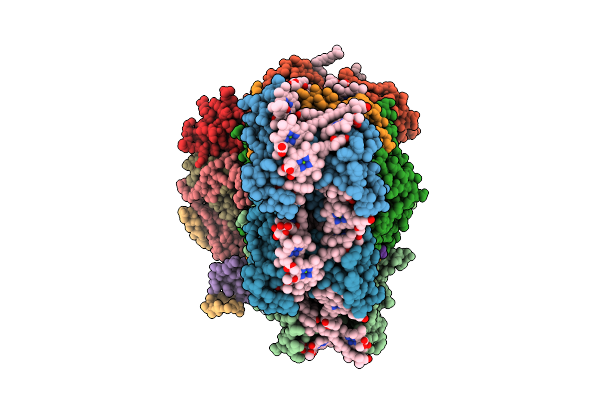 |
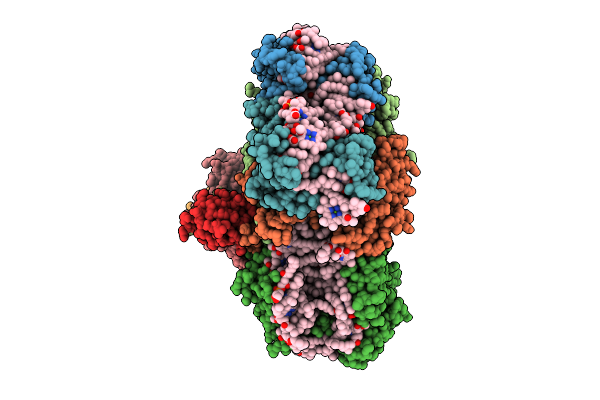
Structure Of The Canthaxanthin Mutant Psi-4Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: A1L1G, A1L1F, XAT, 45D, LHG, CLA, DGD, SQD, PQN, BCR, SF4, LMG |
 |
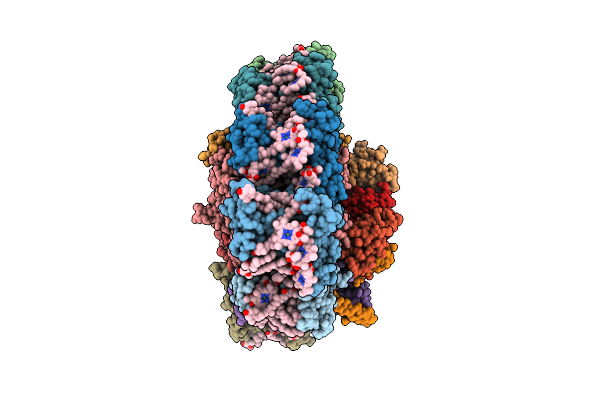
Structure Of The Canthaxanthin Mutant Psi-3Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, DGD, A1L1F, PQN, LHG, BCR, SF4, LMG |
 |
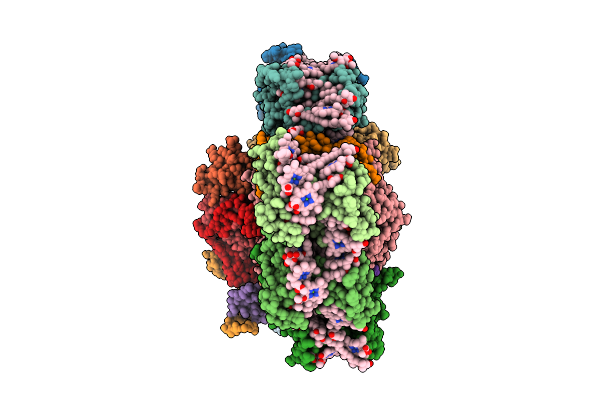
Structure Of The Astaxanthin Mutant Psi-9Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, A1L1F, LHG, DGD, LMG, PQN, BCR, SF4 |
 |
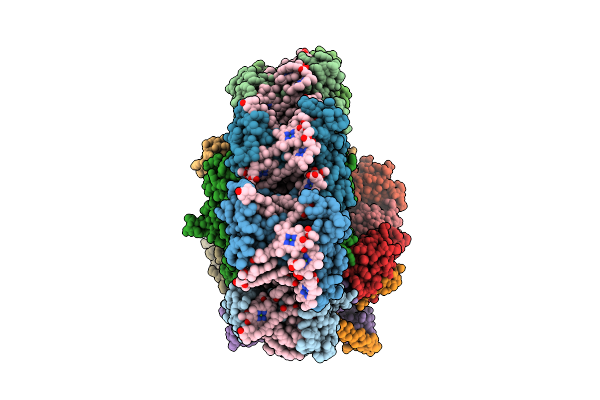
Structure Of The Astaxanthin Mutant Psi-7Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, A1L1F, LHG, DGD, PQN, BCR, SF4, LMG |
 |
Structure Of The Astaxanthin Mutant Psi-5Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, DGD, LMG, A1L1F, PQN, LHG, BCR, SF4 |