Search Count: 23
 |
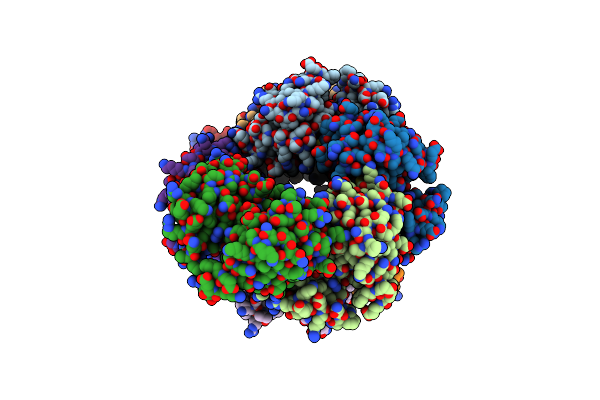
Crystal Structure Of A Slam-Dependent Surface Lipoprotein, Pmslp, In Pasteurella Multocida
Organism: Pasteurella multocida 36950
Method: X-RAY DIFFRACTION Release Date: 2025-04-16 Classification: MEMBRANE PROTEIN Ligands: SE |
 |
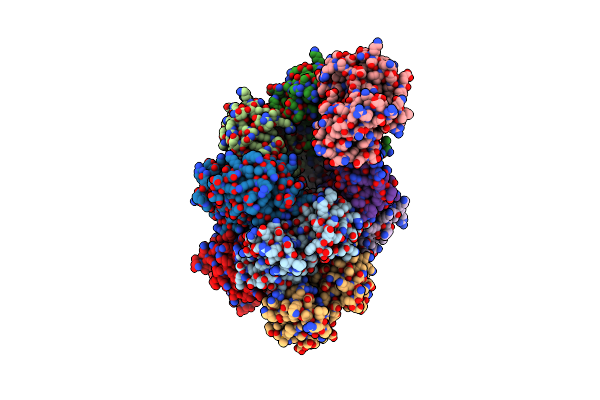
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2024-10-30 Classification: APOPTOSIS Ligands: SE |
 |
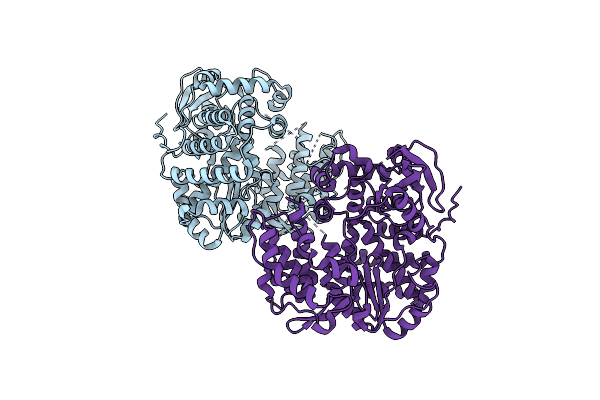
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.32 Å Release Date: 2024-05-15 Classification: APOPTOSIS Ligands: SE |
 |
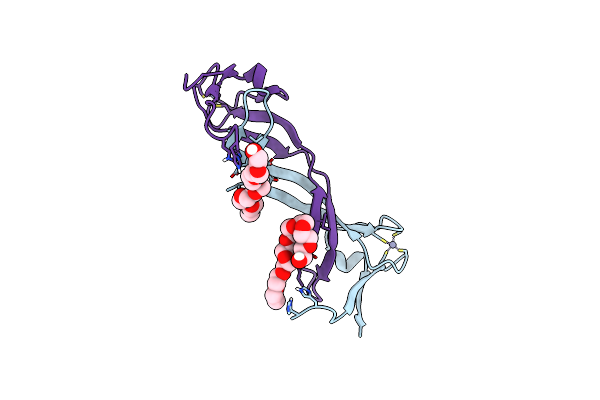
Organism: Pelomicrobium methylotrophicum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-03-27 Classification: OXIDOREDUCTASE Ligands: SEK, CU, GOL, SE, NA |
 |
Organism: Amblyomma virus gxtv108v
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-02-07 Classification: VIRAL PROTEIN Ligands: SE |
 |
Organism: Leptotrichia wadei (strain f0279)
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2023-05-31 Classification: ANTIVIRAL PROTEIN Ligands: PE4, ZN, SE |
 |
Sporosarcina Pasteurii Urease (Spu) Co-Crystallized In The Presence Of An Ebselen-Derivative And Bound To Se Atoms
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Release Date: 2023-04-19 Classification: HYDROLASE Ligands: EDO, SO4, NI, OH, IU9, SE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2023-01-25 Classification: OXIDOREDUCTASE Ligands: ZN, Q8H, ACT, SO4, SE |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-12-14 Classification: HYDROLASE Ligands: SE |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2021-06-02 Classification: VIRAL PROTEIN Ligands: SE |
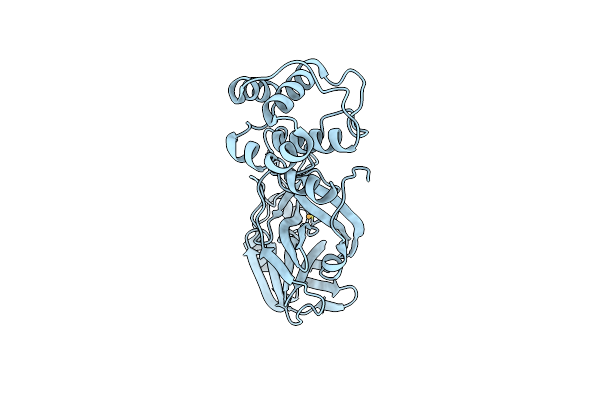 |
Crystal Structure Of Sars-Cov-2 Main Protease Treated With Ebselen Derivative Of Mr6-31-2
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-06-02 Classification: VIRAL PROTEIN Ligands: SE |
 |
Selenium Incorporated Femo-Cofactor Of Nitrogenase From Azotobacter Vinelandii At High Concentration Of Selenium
Organism: Azotobacter vinelandii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-08-14 Classification: OXIDOREDUCTASE Ligands: HCA, ICS, IMD, SE, CA, CLF, MG |
 |
Selenium Incorporated Femo-Cofactor Of Nitrogenase From Azotobacter Vinelandii With Low Concentration Of Selenium
Organism: Azotobacter vinelandii
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-08-14 Classification: OXIDOREDUCTASE Ligands: HCA, ICS, IMD, SE, CLF, MG, CA |
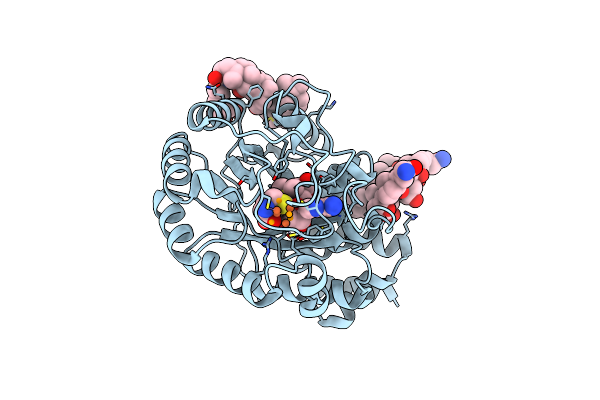 |
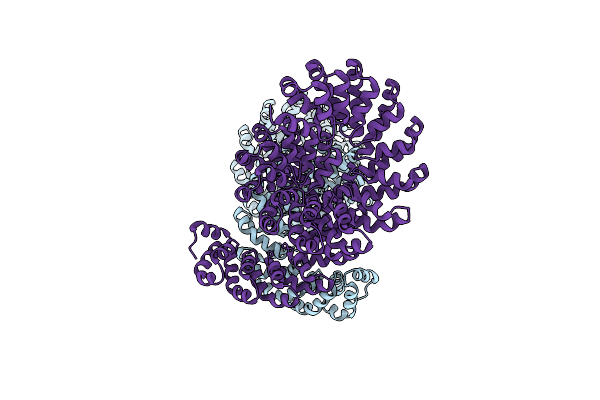
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Release Date: 2016-04-06 Classification: OXIDOREDUCTASE Ligands: SFS, CPS, CL, SAH, 5ZZ, SE |
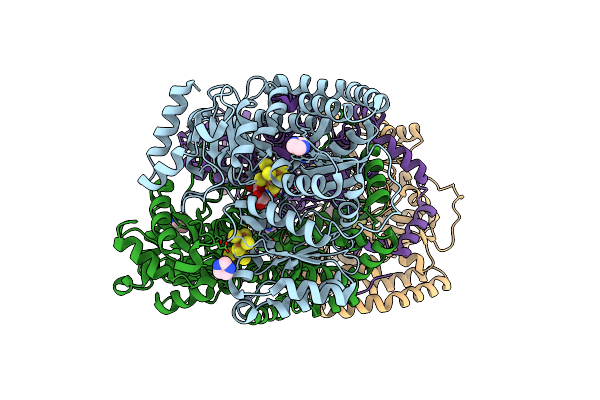 |
Selenium Incorporated Nitrogenase Mofe-Protein (Av1-Se2B) From A. Vinelandii
Organism: Azotobacter vinelandii
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-12-30 Classification: OXIDOREDUCTASE Ligands: HCA, ICG, IMD, CL, CLF, FE2, SE |
 |
Organism: Burkholderia rhizoxinica
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2014-07-09 Classification: TRANSCRIPTION Ligands: SE |
 |
Crystal Structure Of Sulfide:Quinone Oxidoreductase From Acidithiobacillus Ferrooxidans In Complex With Sodium Selenide
Organism: Acidithiobacillus ferrooxidans
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2012-05-16 Classification: OXIDOREDUCTASE Ligands: FAD, LMT, SO4, 6SE, SE |
 |
Crystal Structure Of A Probable Glycerol Dehydrogenase From Sinorhizobium Meliloti 1021
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2011-11-23 Classification: OXIDOREDUCTASE Ligands: GOL, ZN, SE |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:4.30 Å Release Date: 2011-07-27 Classification: TRANSCRIPTION Ligands: SE |
 |
Organism: Eubacterium barkeri
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-06-30 Classification: OXIDOREDUCTASE Ligands: SE, NO3, MG, MOS, MCN, NIO, FAD, FES, CA |

