Search Count: 22
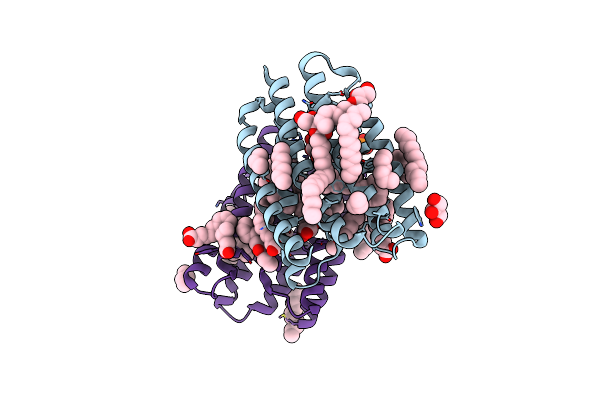 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The Ground State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, GOL, RET, PO4 |
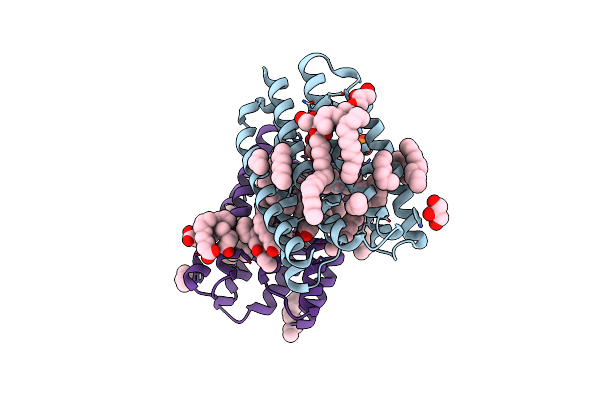 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The P596 State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, GOL, RET, PO4 |
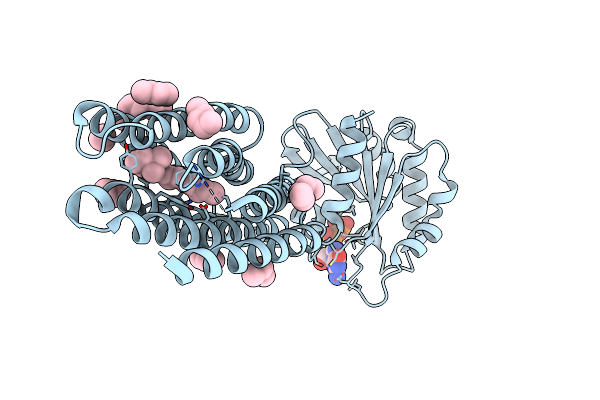 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) - Human Gtpase Arf1 (L8K,Q71L) Chimera; Ground State
Organism: Candidatus actinomarina minuta, Homo sapiens, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: GDP, LFA, RET |
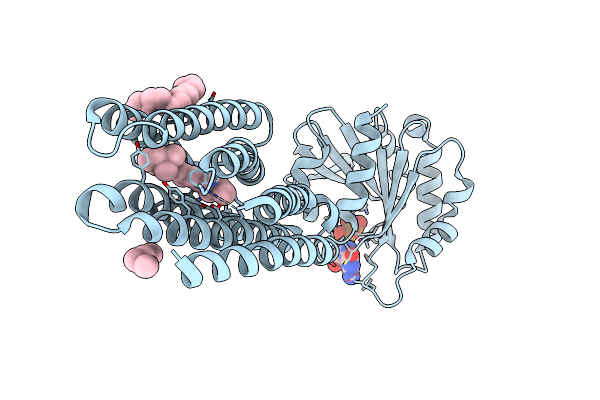 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) - Human Gtpase Arf1 (L8K,Q71L) Chimera; N State
Organism: Candidatus actinomarina minuta, Homo sapiens, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: GDP, LFA, OLA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O* State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O* State, Ph 8.8
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O State Obtained By Cryotrapping
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
True-Atomic Resolution Crystal Structure Of The Closed State Of The Viral Channelrhodopsin Olpvr1
Organism: Organic lake phycodnavirus
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN |
 |
True-Atomic Resolution Crystal Structure Of The Open State Of The Viral Channelrhodopsin Olpvr1
Organism: Organic lake phycodnavirus
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN Ligands: RET, NA, 97N, OLC, LFA, GOL |
 |
Structure Of The Viral Channelrhodopsin Olpvr1 At Low Ph Obtained By Soaking
Organism: Organic lake phycodnavirus
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN Ligands: RET, 97N, LFA, OLC |
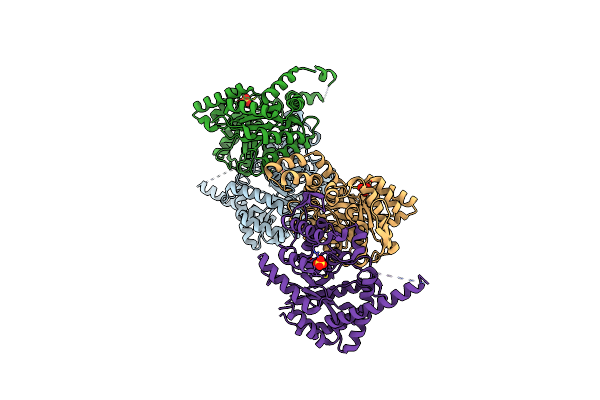 |
Organism: Enhygromyxa salina
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2024-02-07 Classification: LUMINESCENT PROTEIN Ligands: SO4 |
 |
Crystal Structure Of The Microbial Rhodopsin From Sphingomonas Paucimobilis (Spar)
Organism: Sphingomonas paucimobilis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-05-03 Classification: MEMBRANE PROTEIN Ligands: LFA |
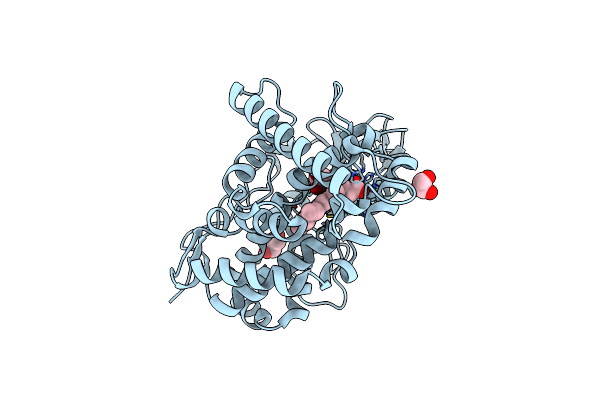 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2023-02-15 Classification: OXIDOREDUCTASE Ligands: HEM, CL, GOL |
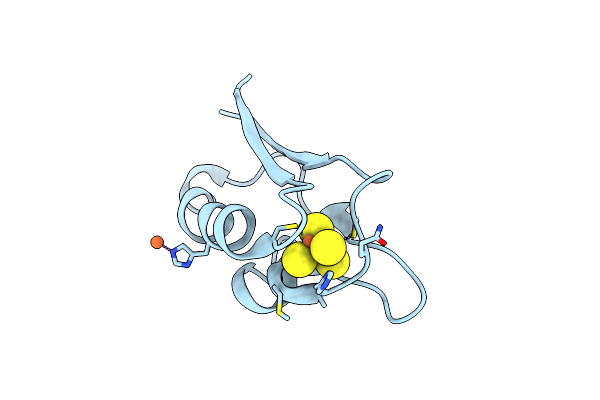 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-02-15 Classification: ELECTRON TRANSPORT Ligands: F3S, FE |
 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-02-15 Classification: OXIDOREDUCTASE Ligands: F3S, HEM, NI |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The M-Like State
Organism: Candidatus actinomarina minuta
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-06-01 Classification: MEMBRANE PROTEIN Ligands: LFA, OLB, OLC, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O State
Organism: Candidatus actinomarina minuta
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2022-06-01 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-04-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA, OLA, HEX, SO4, KR |
 |
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-04-27 Classification: MEMBRANE PROTEIN Ligands: LFA, HEX, NA, KR, RET |
 |
Organism: Candidatus actinomarina minuta
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-04-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA, HEX, KR |

