Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: DNA BINDING PROTEIN Ligands: ZN, ANP, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: DNA BINDING PROTEIN Ligands: ZN, ATP, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: DNA BINDING PROTEIN Ligands: ZN, ANP, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: DNA BINDING PROTEIN Ligands: ZN, ANP, MG |
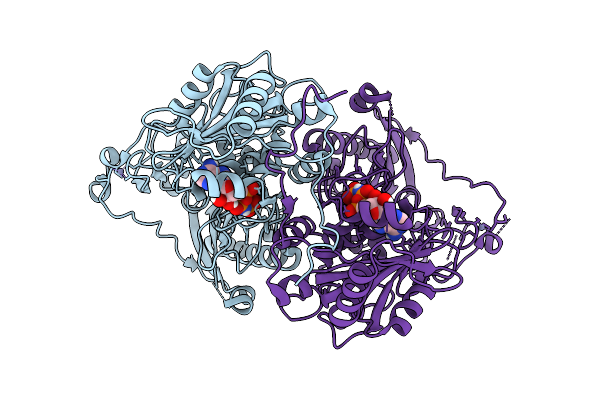 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: DNA BINDING PROTEIN Ligands: ZN, ANP, MG |
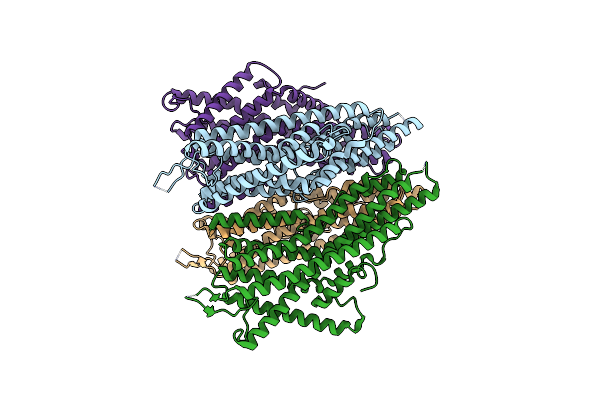 |
Organism: Aedes aegypti, Apocrypta bakeri
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN |
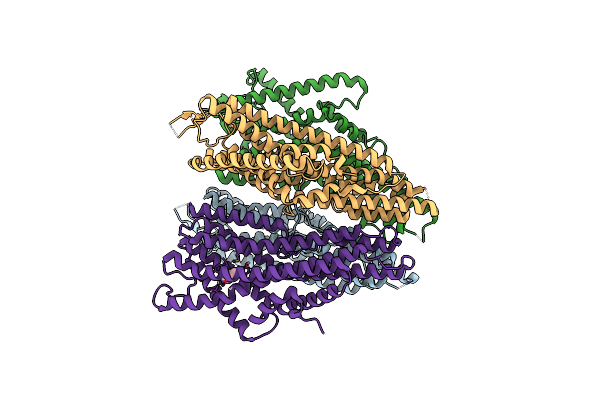 |
Organism: Aedes aegypti, Apocrypta bakeri
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN Ligands: JZ0 |
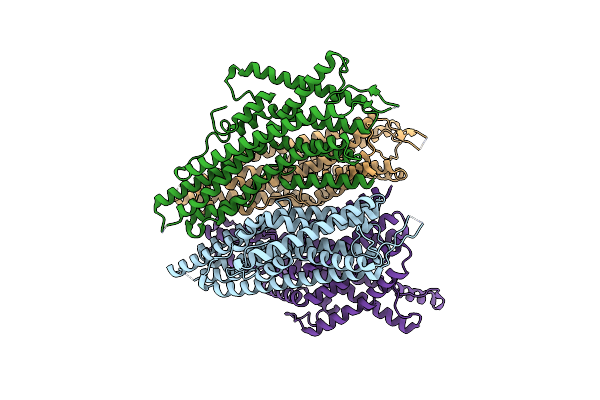 |
Organism: Anopheles gambiae, Apocrypta bakeri
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN |
 |
Organism: Apocrypta bakeri, Anopheles gambiae
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN Ligands: A1AFC |
 |
Crystal Structure Of Co Dehydrogenase Mutant With Increased Affinity For Electron Mediators In High Peg Concentration
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2024-04-17 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, XCC, FE, 1PE |
 |
Crystal Structure Of Co Dehydrogenase Mutant With Increased Affinity For Electron Mediators In Low Peg Concentration
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-04-17 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, XCC, FE, EDO |
 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2024-04-17 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, XCC, S8I, EDO |
 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2024-04-17 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, XCC, PGE, S7L |
 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-04-17 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, XCC, FE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2023-08-23 Classification: UNKNOWN FUNCTION Ligands: NAG, SO4 |
 |
Organism: Human immunodeficiency virus 1
Method: SOLUTION NMR Release Date: 2022-10-26 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2022-05-18 Classification: OXIDOREDUCTASE Ligands: PC1, 3PE, FES, CDL, FMN, SF4, CU, HEA, MG, ZN, NAP, HEM, HEC, UQ2 |
 |
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2022-05-18 Classification: OXIDOREDUCTASE Ligands: FES, 3PE, CDL, PC1, FMN, SF4, ZN, NAP, HEM, HEC, HEA, CU, MG |
 |
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2022-05-18 Classification: OXIDOREDUCTASE Ligands: FES, CDL, 3PE, PC1, FMN, SF4, ZN, NAP, HEM, HEC, HEA, CU, MG |
 |
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2022-05-18 Classification: OXIDOREDUCTASE Ligands: 3PE, CDL, FMN, SF4, FES, ZN, PC1, NAP |