Search Count: 37
 |
Organism: Paenibacillus popilliae
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: TOXIN Ligands: MG |
 |
Organism: Paenibacillus popilliae
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: TOXIN |
 |
Organism: Oryza sativa
Method: X-RAY DIFFRACTION Resolution:3.16 Å Release Date: 2023-12-06 Classification: OXIDOREDUCTASE |
 |
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:3.22 Å Release Date: 2021-05-05 Classification: TOXIN Ligands: CA, SO4 |
 |
Organism: Brevibacillus laterosporus
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2021-05-05 Classification: TOXIN Ligands: SM, ACT, EDO, NA |
 |
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2021-03-17 Classification: TOXIN |
 |
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-02-26 Classification: OXIDOREDUCTASE Ligands: FES |
 |
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2019-06-19 Classification: TOXIN |
 |
Crystal Structure Of A Bacillus Thuringiensis Cry1B.867 Tryptic Core Variant
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-06-19 Classification: TOXIN |
 |
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2018-09-12 Classification: TOXIN |
 |
Organism: Sphingobium herbicidovorans (strain atcc 700291 / dsm 11019 / nbrc 16415 / mh)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-08-15 Classification: OXIDOREDUCTASE Ligands: CO, AKG |
 |
Organism: Sphingobium herbicidovorans (strain atcc 700291 / dsm 11019 / nbrc 16415 / mh)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-08-15 Classification: OXIDOREDUCTASE |
 |
Organism: Sphingobium herbicidovorans
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2018-08-15 Classification: OXIDOREDUCTASE Ligands: FTV, CO, AKG, CL |
 |
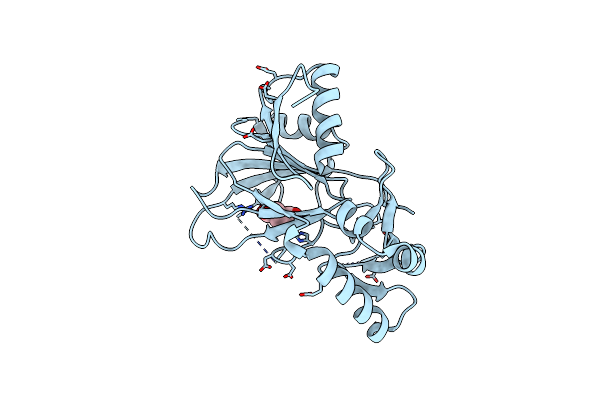
Organism: Sphingobium herbicidovorans
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2018-08-15 Classification: OXIDOREDUCTASE Ligands: CO, AKG, CFA |
 |
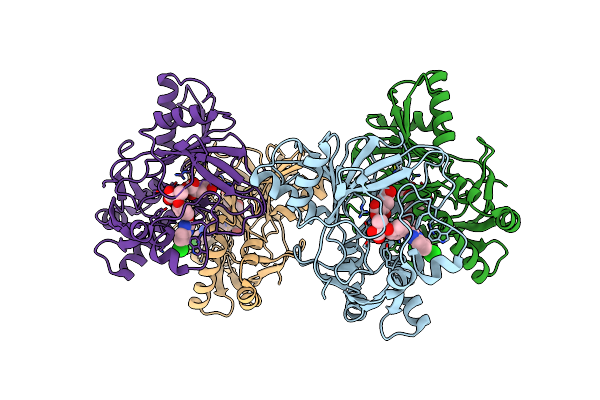
Organism: Sphingobium herbicidovorans
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2018-08-15 Classification: OXIDOREDUCTASE Ligands: CO, AKG |
 |
Organism: Sphingobium herbicidovorans
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2018-08-15 Classification: OXIDOREDUCTASE Ligands: CO, FTJ, AKG, CL |
 |
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2016-07-20 Classification: TOXIN |
 |
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2015-10-28 Classification: TOXIN Ligands: ZN, CA |
 |
Structure Of The Full-Length Insecticidal Protein Cry1Ac Reveals Intriguing Details Of Toxin Packaging Into In Vivo Formed Crystals
Organism: Bacillus thuringiensis serovar kurstaki str. hd73
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2014-09-17 Classification: TOXIN Ligands: K, CA |
 |
Organism: Hordeum vulgare
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2012-07-18 Classification: TRANSFERASE Ligands: DCS |

