Search Count: 17
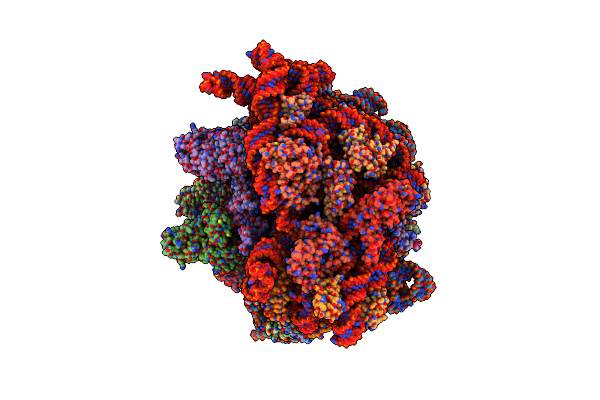 |
Cryo-Em Structure Of An E. Coli Non-Rotated Ribosome Termination Complex Bound With Aporf3, Rf1, P- And E-Site Trnaphe (Composite State I-B)
Organism: Escherichia coli, Escherichia coli k-12, Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: RIBOSOME Ligands: MG, ZN |
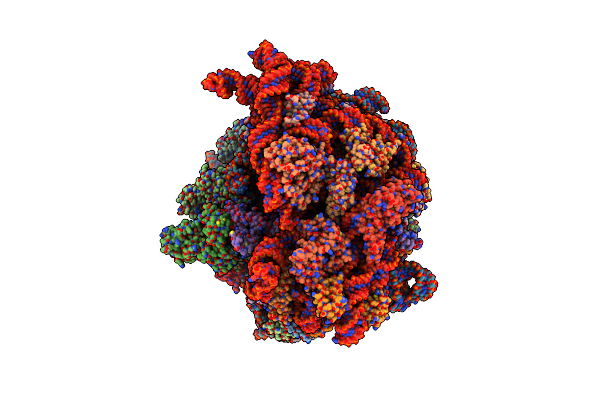 |
Cryo-Em Structure Of An E. Coli Non-Rotated Ribosome Termination Complex Bound With Rf1, P- And E-Site Trnaphe (State I-A)
Organism: Escherichia coli, Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: RIBOSOME Ligands: MG, ZN |
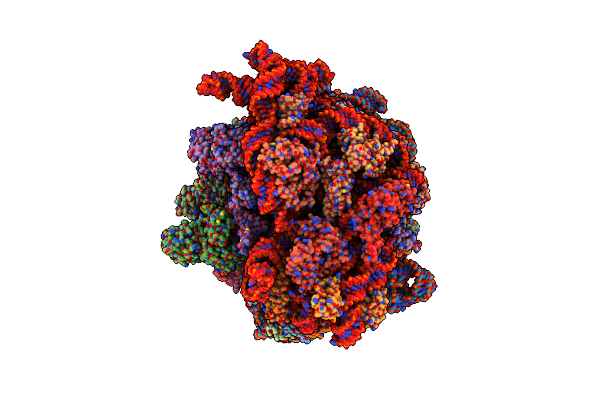 |
Cryo-Em Structure Of An E. Coli Non-Rotated Ribosome Termination Complex Bound With Rf3-Gdpcp, Rf1, P- And E-Site Trnaphe (Composite State Ii-A)
Organism: Escherichia coli, Escherichia coli k-12, Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: RIBOSOME Ligands: MG, GCP, ZN |
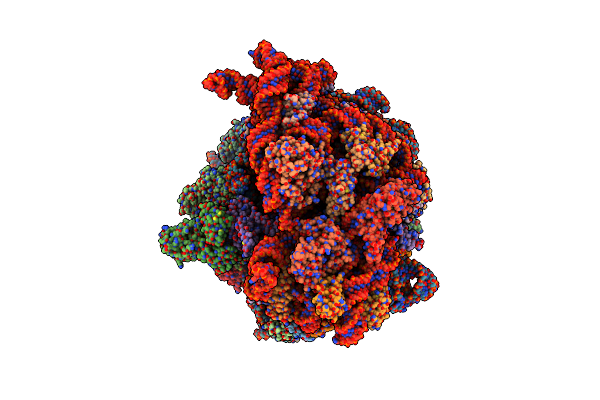 |
Cryo-Em Structure Of An E. Coli Non-Rotated Ribosome Termination Complex Bound With Rf1, P- And E-Site Trnaphe (State Ii-D)
Organism: Escherichia coli, Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: RIBOSOME Ligands: MG, ZN |
 |
Cryo-Em Structure Of An E. Coli Rotated Ribosome Bound With Rf3-Gdpcp And P/E-Trnaphe (Composite State Ii-B)
Organism: Escherichia coli, Escherichia coli k-12, Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: RIBOSOME Ligands: MG, GCP, ZN |
 |
Cryo-Em Structure Of An E. Coli Rotated Ribosome Bound With Rf3-Gdpcp And P/E-Trnaphe (Composite State Ii-C)
Organism: Escherichia coli, Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: RIBOSOME Ligands: MG, GCP, ZN |
 |
Cryo-Em Structure Of An E. Coli Rotated Ribosome Complex Bound With Rf3-Ppgpp And P/E-Trnaphe (Composite State I-C)
Organism: Escherichia coli, Escherichia coli k-12, Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: RIBOSOME/RNA Ligands: MG, G4P, ZN |
 |
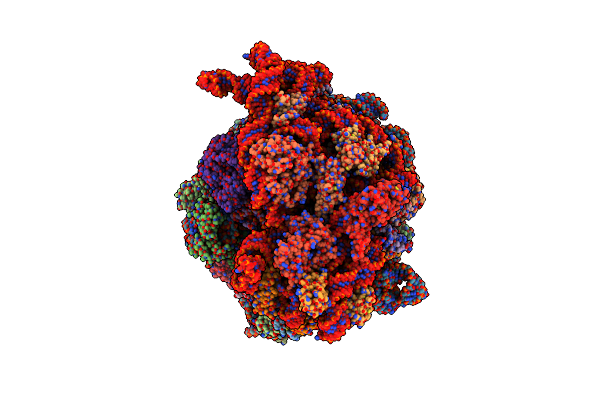
Structure Of The Escherichia Coli 70S Ribosome In Complex With Ef-Tu And Ile-Trnaile(Lau) Bound To The Cognate Aua Codon (Structure I)
Organism: Escherichia coli, Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: RIBOSOME Ligands: MG, K, ILE, GCP, ZN |
 |
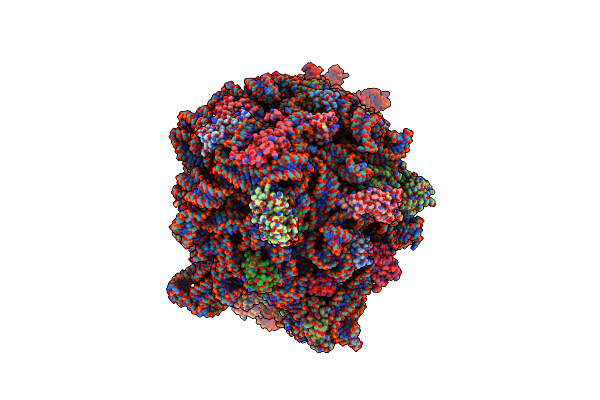
Structure Of The Escherichia Coli 70S Ribosome In Complex With Ef-Tu And Ile-Trnaile(Lau) Bound To The Near-Cognate Aug Codon (Structure Ii)
Organism: Escherichia coli, Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: RIBOSOME Ligands: PAR, MG, ILE, GCP, ZN |
 |
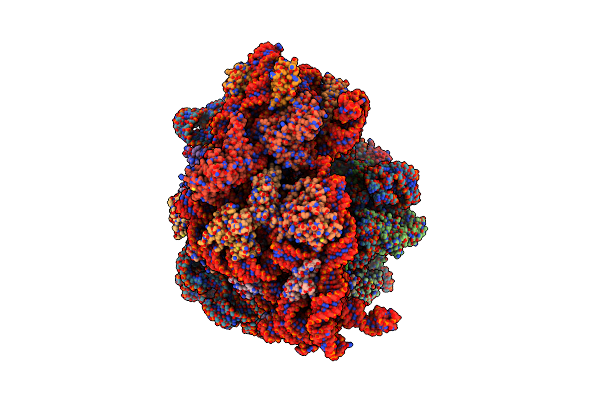
Structure Of The Escherichia Coli 70S Ribosome In Complex With A-Site Trnaile(Lau) Bound To The Cognate Aua Codon (Structure Iii)
Organism: Escherichia coli, Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: RIBOSOME Ligands: PAR, MG, K, ILE, ZN |
 |
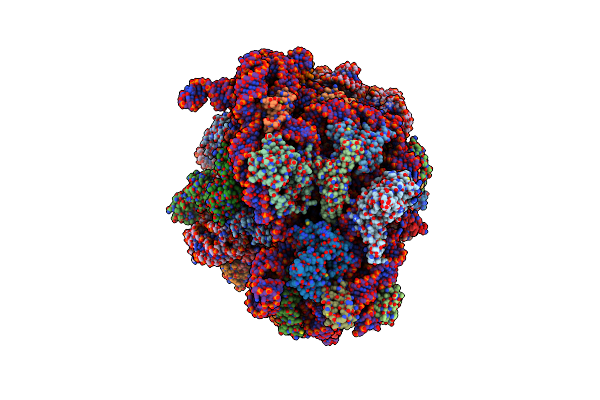
Structure Of The Escherichia Coli 70S Ribosome In Complex With P-Site Trnaile(Lau) Bound To The Cognate Aua Codon (Structure Iv)
Organism: Escherichia coli, Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: RIBOSOME Ligands: MG, ILE, ZN |
 |
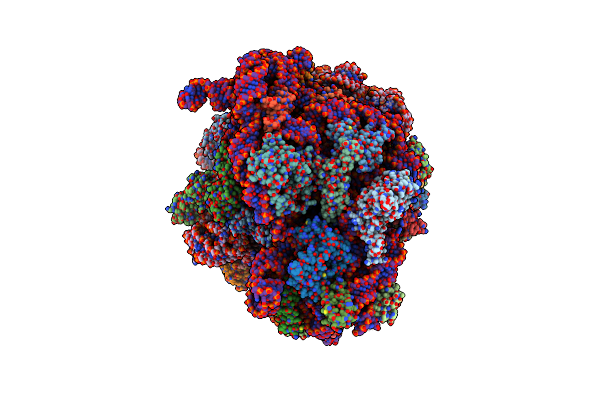
Cryo-Em Structure Of P. Urativorans 70S Ribosome In Complex With Hibernation Factors Balon And Raia (Structure 1).
Organism: Psychrobacter urativorans
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: RIBOSOME Ligands: MG |
 |
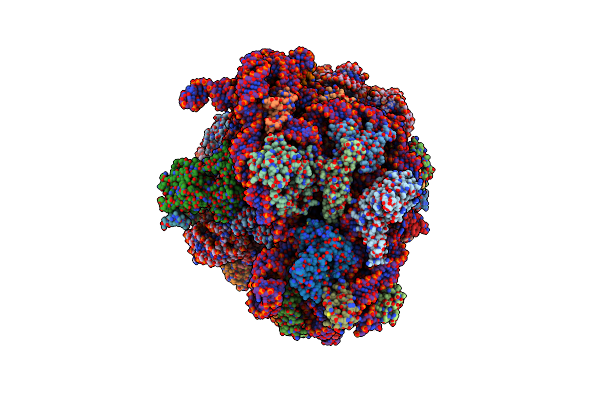
Cryo-Em Structure Of P. Urativorans 70S Ribosome In Complex With Hibernation Factor Balon, Mrna And P-Site Trna (Structure 2).
Organism: Psychrobacter urativorans
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: RIBOSOME |
 |
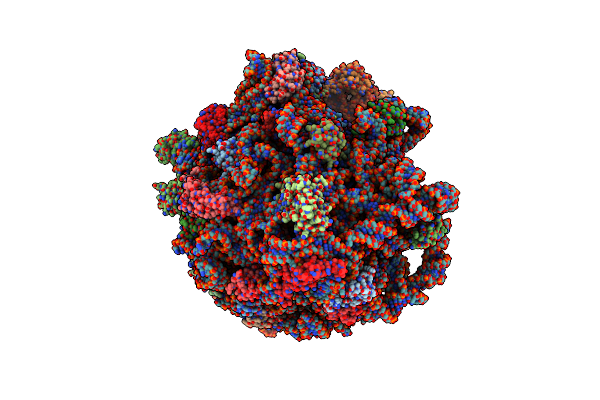
Cryo-Em Structure Of P. Urativorans 70S Ribosome In Complex With Hibernation Factor Balon And Ef-Tu(Gdp) (Structure 3).
Organism: Psychrobacter urativorans
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: RIBOSOME Ligands: GDP, MG |
 |
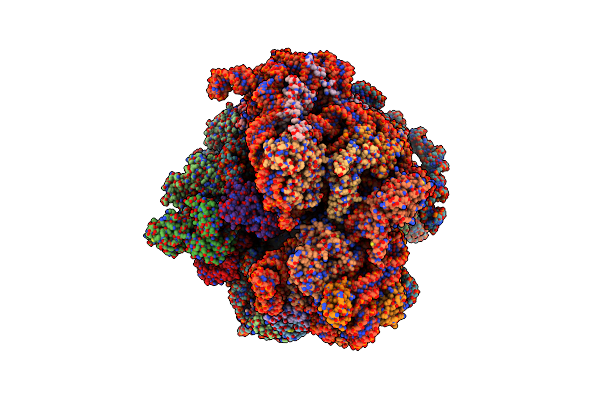
Cryo-Em Structure Of The Mycobacterium Smegmatis 70S Ribosome In Complex With Hibernation Factor Msmeg1130 (Balon) (Structure 4)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: RIBOSOME Ligands: ZN |
 |
Cryo-Em Structure Of The Mycobacterium Smegmatis 70S Ribosome In Complex With Hibernation Factor Rv2629 (Balon) (Structure 5)
Organism: Mycobacterium tuberculosis h37rv, Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: RIBOSOME Ligands: ZN |
 |
Cryo-Em Structure Of The Mycobacterium Smegmatis 70S Ribosome In Complex With Hibernation Factor Msmeg1130 (Balon) And Msmegef-Tu(Gdp) (Composite Structure 6)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: RIBOSOME Ligands: GDP, ZN |

