Search Count: 44
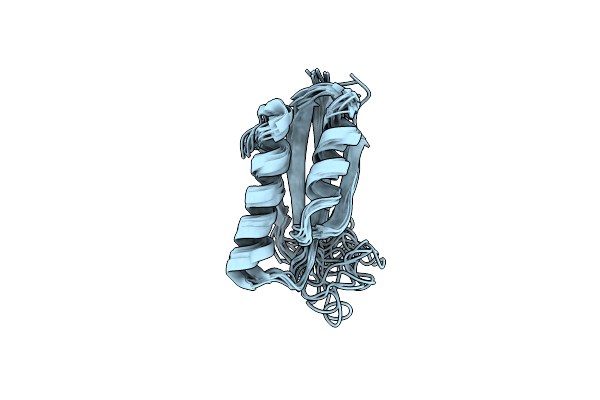 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2023-01-11 Classification: STRUCTURAL PROTEIN |
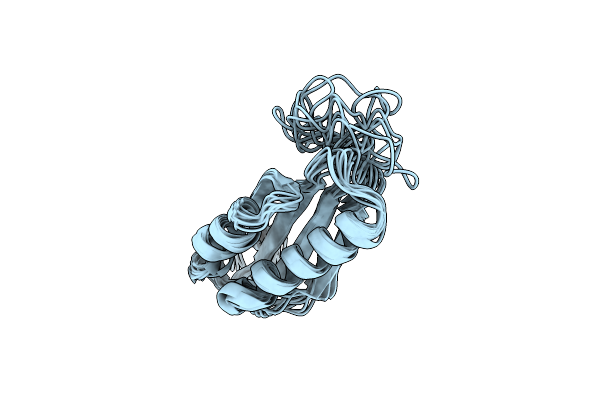 |
Rules For Designing Protein Fold Switches And Their Implications For The Folding Code
Organism: Thermus thermophilus
Method: SOLUTION NMR Release Date: 2022-05-18 Classification: DE NOVO PROTEIN |
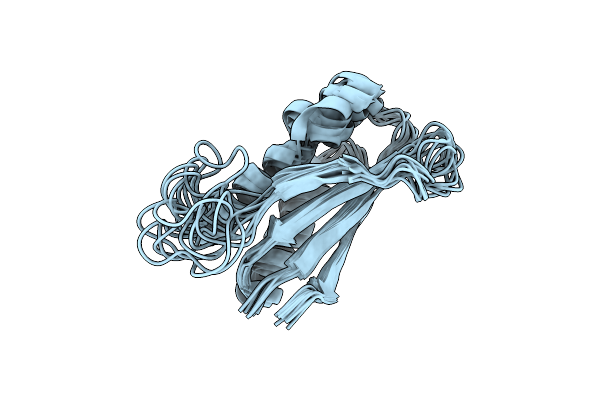 |
Rules For Designing Protein Fold Switches And Their Implications For The Folding Code
Organism: Thermus thermophilus
Method: SOLUTION NMR Release Date: 2022-05-18 Classification: DE NOVO PROTEIN |
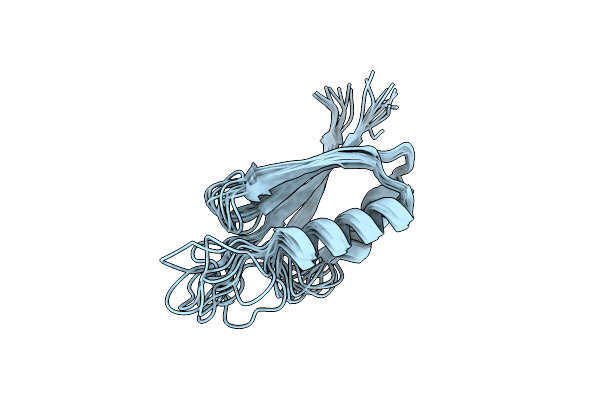 |
Rules For Designing Protein Fold Switches And Their Implications For The Folding Code
Organism: Thermus thermophilus
Method: SOLUTION NMR Release Date: 2022-05-18 Classification: DE NOVO PROTEIN |
 |
Rules For Designing Protein Fold Switches And Their Implications For The Folding Code
Organism: Thermus thermophilus
Method: SOLUTION NMR Release Date: 2022-05-18 Classification: DE NOVO PROTEIN |
 |
Organism: Bacillus amyloliquefaciens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-09-16 Classification: HYDROLASE Ligands: K, GOL, SCN |
 |
Imidazole-Triggered Ras-Specific Subtilisin Subt_Bacam Complexed With Ysam Peptide
Organism: Bacillus amyloliquefaciens, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2020-09-16 Classification: HYDROLASE |
 |
Imidazole-Triggered Ras-Specific Subtilisin Subt_Bacam Complexed With The Peptide Eeysam
Organism: Bacillus amyloliquefaciens, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2020-09-16 Classification: HYDROLASE |
 |
Organism: Bacillus amyloliquefaciens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-09-16 Classification: HYDROLASE |
 |
Organism: Thermosynechococcus elongatus (strain bp-1)
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2019-01-02 Classification: PROTEIN BINDING Ligands: CO, SO4, CL, SCN |
 |
Organism: Homo sapiens, Human respiratory syncytial virus a (strain a2)
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2017-05-17 Classification: Viral Protein/Immune System Ligands: CL |
 |
Organism: Human respiratory syncytial virus 9320, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:4.30 Å Release Date: 2017-05-17 Classification: Viral Protein/Immune System Ligands: SO4 |
 |
Organism: Human respiratory syncytial virus 9320
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2017-05-17 Classification: VIRAL PROTEIN Ligands: SO4 |
 |
Organism: Streptococcus dysgalactiae
Method: SOLUTION NMR Release Date: 2015-04-08 Classification: PROTEIN BINDING |
 |
Organism: Plasmodium falciparum
Method: SOLUTION NMR Release Date: 2012-10-03 Classification: HYDROLASE |
 |
Organism: Artificial gene
Method: SOLUTION NMR Release Date: 2012-02-29 Classification: DE NOVO PROTEIN |
 |
Organism: Artificial gene
Method: SOLUTION NMR Release Date: 2012-02-29 Classification: DE NOVO PROTEIN |
 |
Organism: Artificial gene
Method: SOLUTION NMR Release Date: 2012-02-29 Classification: DE NOVO PROTEIN |
 |
Organism: Artificial gene
Method: SOLUTION NMR Release Date: 2012-02-29 Classification: DE NOVO PROTEIN |
 |
Nmr Structures Of Ga95 And Gb95, Two Designed Proteins With 95% Sequence Identity But Different Folds And Functions
|

