Search Count: 28
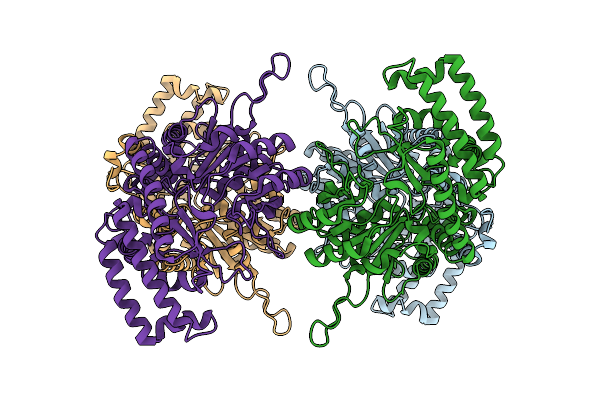 |
Cryo-Em Structure Of Human Serine Hydroxymethyltransferase, Isoform 2 (Shmt2)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-09-20 Classification: TRANSFERASE |
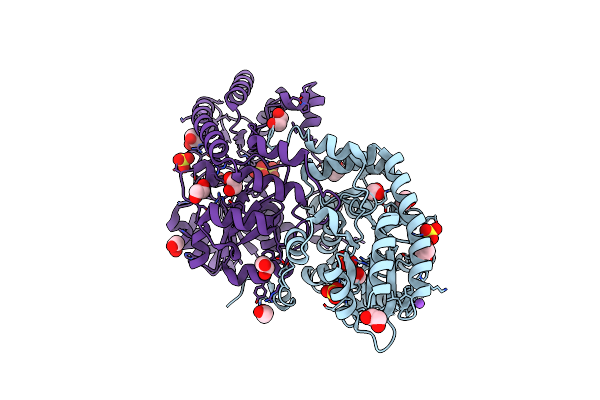 |
Crystal Structure Of Medicago Truncatula Histidinol-Phosphate Aminotransferase (Hisn6) In The Open State
Organism: Medicago truncatula
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2023-03-22 Classification: TRANSFERASE Ligands: SO4, EDO, NA |
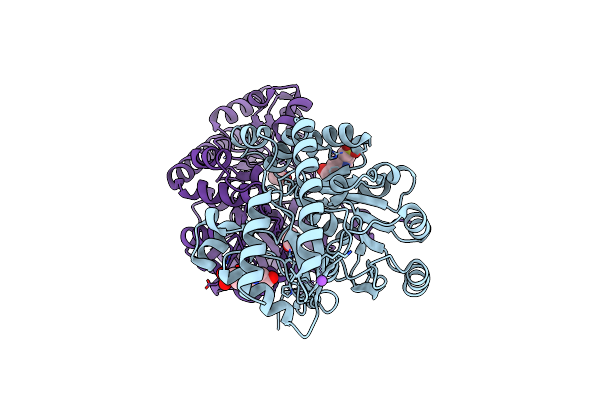 |
Crystal Structure Of Medicago Truncatula Histidinol-Phosphate Aminotransferase (Hisn6) In The Closed State
Organism: Medicago truncatula
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2023-03-22 Classification: TRANSFERASE Ligands: ACT, NA, EPE |
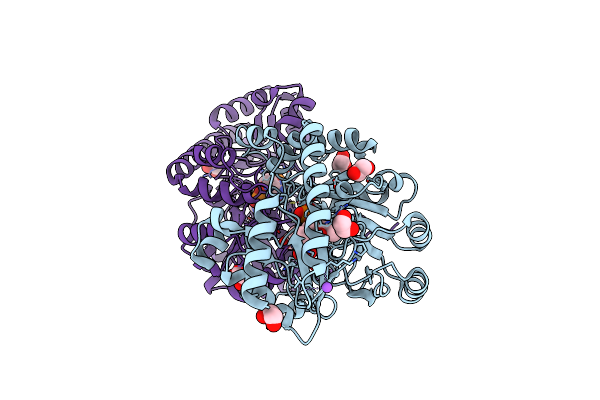 |
Crystal Structure Of Medicago Truncatula Histidinol-Phosphate Aminotransferase (Hisn6) In Complex With Histidinol-Phosphate
Organism: Medicago truncatula
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2023-03-22 Classification: TRANSFERASE Ligands: QNX, EDO, NA |
 |
Crystal Structure Of Medicago Truncatula Histidinol-Phosphate Aminotransferase (Hisn6) In Apo Form
Organism: Medicago truncatula
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2023-03-22 Classification: TRANSFERASE Ligands: SO4, EDO, NA |
 |
Psychrophilic Aromatic Amino Acids Aminotransferase From Psychrobacter Sp. B6 Cocrystalized With Substrate Analog - L-(-)-3-Phenyllactic Acid
Organism: Psychrobacter sp. b6
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-07-14 Classification: TRANSFERASE Ligands: MG, PLP, NO3, HFA |
 |
Psychrophilic Aromatic Amino Acids Aminotransferase From Psychrobacter Sp. B6 Cocrystalized With Substrate Analog - L-P-Hydroxyphenyllactic Acid
Organism: Psychrobacter sp. b6
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2021-07-14 Classification: TRANSFERASE Ligands: MG, TYF, NO3, PLP |
 |
Psychrophilic Aromatic Amino Acids Aminotransferase From Psychrobacter Sp. B6 Cocrystalized With Substrate Analog - L-Indole-3-Lactic Acid
Organism: Psychrobacter sp. b6
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2021-07-14 Classification: TRANSFERASE Ligands: MG, NO3, PLP, 3IL |
 |
Psychrophilic Aromatic Amino Acids Aminotransferase From Psychrobacter Sp. B6 Cocrystalized With Substrate Analog - Malic Acid
Organism: Psychrobacter sp. b6
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2020-11-18 Classification: TRANSFERASE Ligands: PLP, LMR |
 |
Organism: Arthrobacter sp. 32cb
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-08-05 Classification: HYDROLASE |
 |
Cold-Adapted Beta-D-Galactosidase From Arthrobacter Sp. 32Cb Mutant E517Q In Complex With Galactose
Organism: Arthrobacter sp. 32cb
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-08-05 Classification: HYDROLASE |
 |
Cold-Adapted Beta-D-Galactosidase From Arthrobacter Sp. 32Cb Mutant E517Q In Complex With Lactulose
Organism: Arthrobacter sp. 32cb
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-08-05 Classification: HYDROLASE |
 |
Cold-Adapted Beta-D-Galactosidase From Arthrobacter Sp. 32Cb Mutant E441Q In Complex With Galactose
Organism: Arthrobacter sp. 32cb
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-08-05 Classification: HYDROLASE Ligands: GAL, NA, ACT, MLI, FMT, LMR |
 |
Cold-Adapted Beta-D-Galactosidase From Arthrobacter Sp. 32Cb Mutant E441Q In Complex With Lactulose
Organism: Arthrobacter sp. 32cb
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2020-08-05 Classification: HYDROLASE Ligands: GAL, ACT, NA |
 |
Cold-Adapted Beta-D-Galactosidase From Arthrobacter Sp. 32Cb Mutant E441Q In Complex With Saccharose
Organism: Arthrobacter sp. 32cb
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-08-05 Classification: HYDROLASE Ligands: ACT, NA, MLI |
 |
Organism: Arthrobacter sp. 32cb
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-08-05 Classification: HYDROLASE Ligands: MLI |
 |
Cold-Adapted Beta-D-Galactosidase From Arthrobacter Sp. 32Cb Mutant D207A In Complex With Galactose
Organism: Arthrobacter sp. 32cb
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2020-08-05 Classification: HYDROLASE Ligands: GAL |
 |
Cold-Adapted Beta-D-Galactosidase From Arthrobacter Sp. 32Cb Mutant D207A In Complex With Saccharose
Organism: Arthrobacter sp. 32cb
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2020-08-05 Classification: HYDROLASE |
 |
Crystal Structure Of Human Prolidase S202F Variant Expressed In The Presence Of Chaperones
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2020-07-15 Classification: HYDROLASE Ligands: MN, GOL, MH2, GLY, PRO |
 |
Crystal Structure Of Human Prolidase G278N Variant Expressed In The Presence Of Chaperones
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-07-15 Classification: HYDROLASE Ligands: MN, NA, GOL, GLY, PRO |

