Search Count: 8
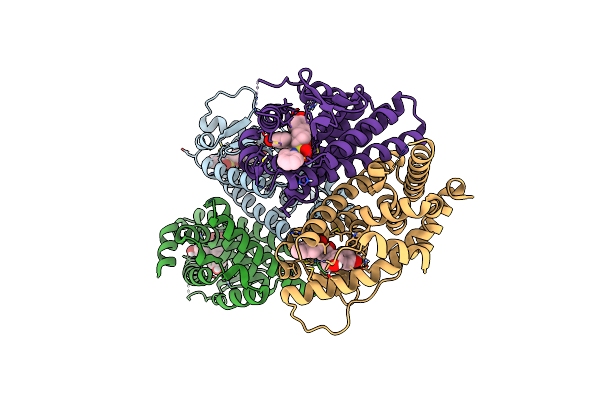 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-411
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHV |
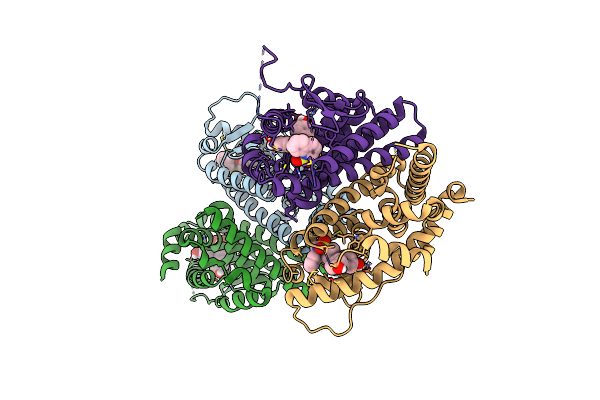 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-410
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHU |
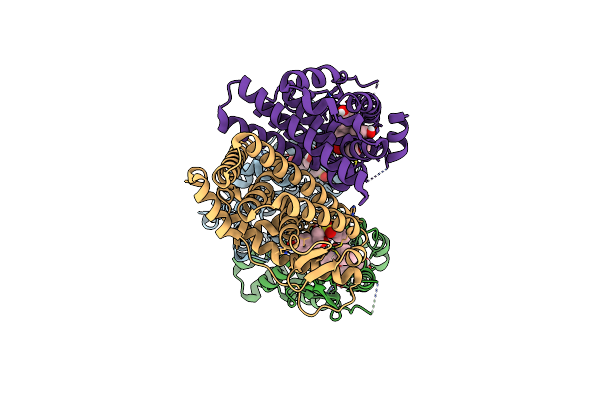 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-400
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHO |
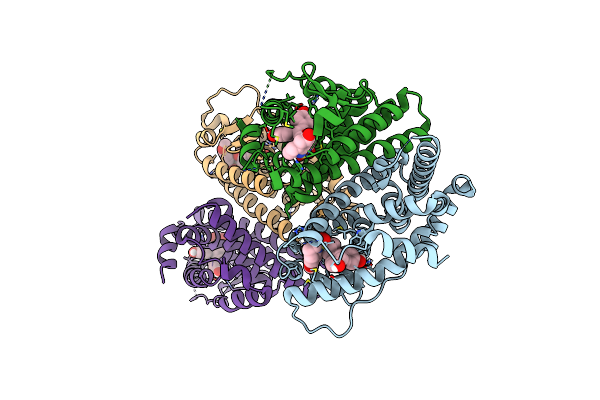 |
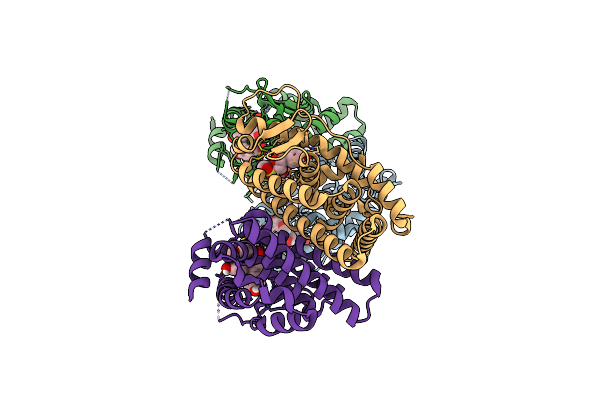
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-409
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHS |
 |
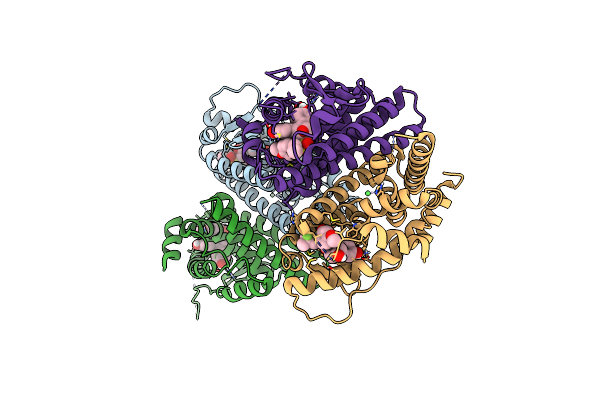
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-403
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHW |
 |
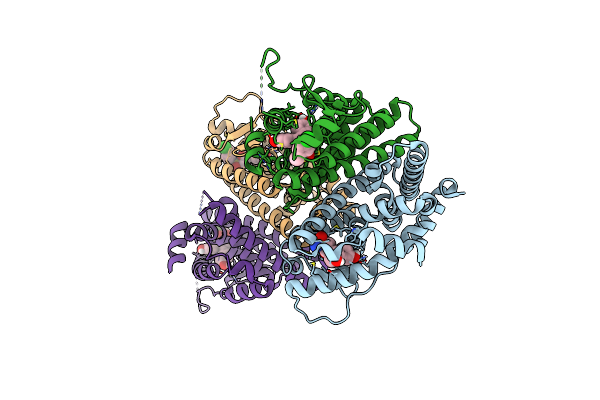
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-406
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHX, NI |
 |
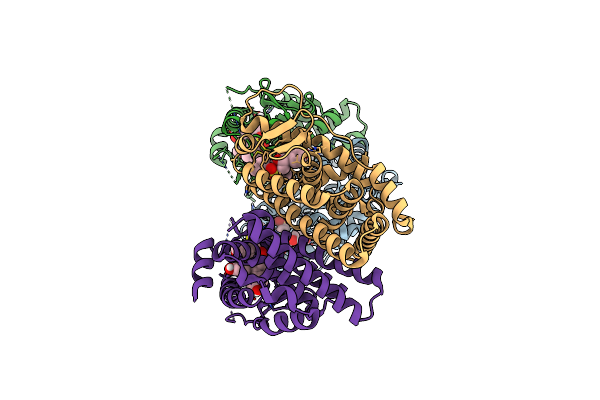
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-1154
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: OBT |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-402
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHY, A1AHZ |

