Search Count: 46
 |
The Histone Fold Domain Heterodimer Of Oocyst Rupture Proteins 1 And 2 From Plasmodium Berghei
Organism: Plasmodium berghei
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2026-01-21 Classification: UNKNOWN FUNCTION Ligands: SO4, EDO |
 |
The Structure Of The Dna-Binding Domain Of Nuclear Factor 1 X Bound To Nfi Consensus Dna Sequence
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Resolution:3.86 Å Release Date: 2025-09-17 Classification: TRANSCRIPTION Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2025-07-09 Classification: IMMUNE SYSTEM Ligands: PEG, PO4, PGE, EDO, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2024-07-10 Classification: BIOSYNTHETIC PROTEIN Ligands: SF4, SAM, MET, SO4, GOL, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2024-07-10 Classification: BIOSYNTHETIC PROTEIN Ligands: SF4, MET |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-411
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHV |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-410
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHU |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-400
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHO |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-409
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHS |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-403
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHW |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-406
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHX, NI |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-1154
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: OBT |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-402
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHY, A1AHZ |
 |
Structural Characterization Of Phox2B And Its Dna Interactions Shed Lights Into The Molecular Basis Of The + 7Ala Variant Pathogenicity In Cchs
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2024-05-15 Classification: PROTEIN BINDING |
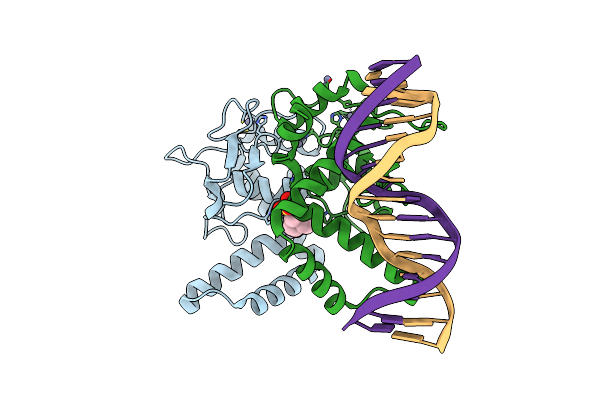 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-01-18 Classification: DNA BINDING PROTEIN Ligands: ZN, EPE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2023-01-18 Classification: DNA BINDING PROTEIN Ligands: ZN, EPE |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2021-03-31 Classification: IMMUNE SYSTEM Ligands: CAC, IMD, NA |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-04-01 Classification: TRANSFERASE |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2020-04-01 Classification: TRANSFERASE Ligands: JFX, NA, GOL, IPA |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2020-04-01 Classification: TRANSFERASE |

