Search Count: 74
 |
Impacts Of Ribosomal Rna Sequence Variation On Gene Expression And Phenotype: Cryo-Em Structure Of The Rrsb Ribosome (Bbb-70S)
Organism: Escherichia coli k-12, Escherichia coli bl21
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: RIBOSOME Ligands: PUT, SPD, MG, K, ZN, ATP, FME |
 |
Impacts Of Ribosomal Rna Sequence Variation On Gene Expression And Phenotype: Cryo-Em Structure Of The Rrsh Ribosome (Hbb-70S)
Organism: Escherichia coli k-12, Escherichia coli bl21
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: RIBOSOME Ligands: PUT, SPD, MG, K, ZN, ATP, FME |
 |
Mrna Decoding In Human Is Kinetically And Structurally Distinct From Bacteria (Ic State)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: RIBOSOME Ligands: SPD, PUT, K, MG, ANM, 3H3, ZN, MET |
 |
Mrna Decoding In Human Is Kinetically And Structurally Distinct From Bacteria (Ga State)
Organism: Escherichia coli, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: RIBOSOME Ligands: SPD, K, MG, ANM, PUT, 3H3, ZN, PHE, MET, GSP, ZIY |
 |
Mrna Decoding In Human Is Kinetically And Structurally Distinct From Bacteria (Cr State)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: RIBOSOME Ligands: SPD, PUT, MG, ANM, 3H3, K, ZN, PHE, MET, GSP, ZIY |
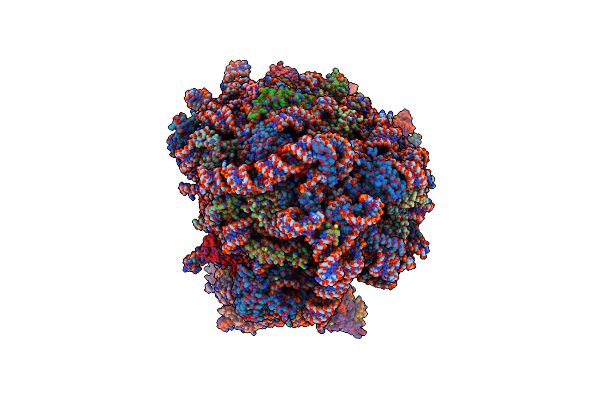 |
Mrna Decoding In Human Is Kinetically And Structurally Distinct From Bacteria (Ac State)
Organism: Homo sapiens, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: RIBOSOME Ligands: PUT, K, MG, ANM, SPD, 3H3, ZN, MET |
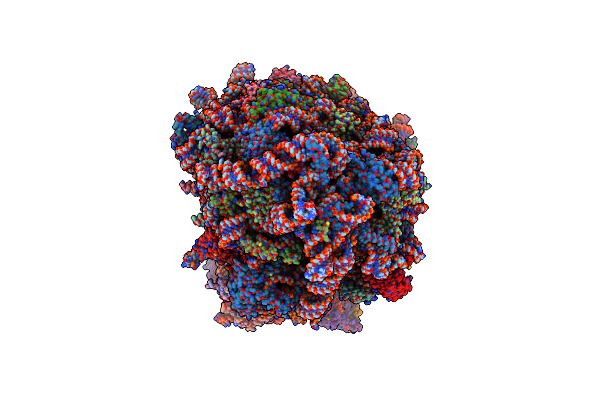 |
Mrna Decoding In Human Is Kinetically And Structurally Distinct From Bacteria (Ga State 2)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: RIBOSOME Ligands: SPD, K, MG, PUT, 3HE, HMT, ZN, PHE, MET, GSP, YRB |
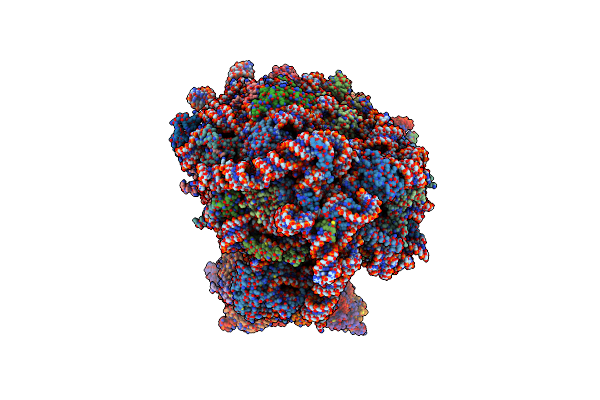 |
Mrna Decoding In Human Is Kinetically And Structurally Distinct From Bacteria (Consensus Lsu Focused Refined Structure)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: RIBOSOME Ligands: SPD, PUT, K, MG, ANM, 3H3, TRS, ZN, MET |
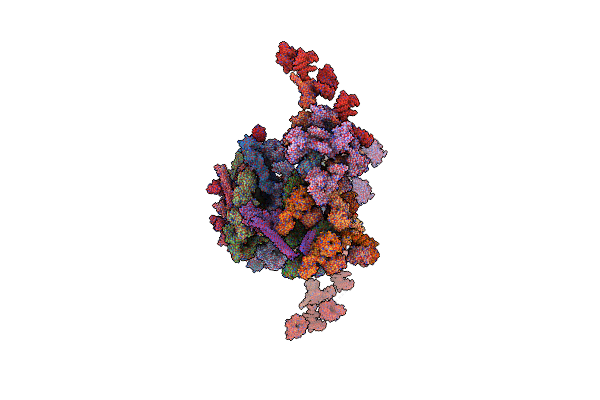 |
Composite Structure Of The Human Nuclear Pore Complex (Npc) Cytoplasmic Face Generated With A 12A Cryo-Et Map Of The Purified Hela Cell Npc
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-06-22 Classification: TRANSPORT PROTEIN |
 |
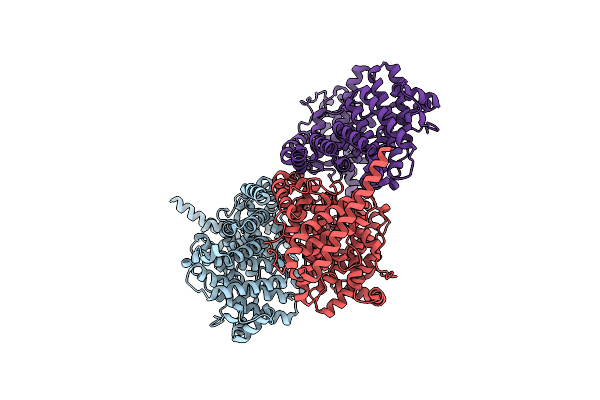
Crystal Structure Of The N-Terminal Domain Of Nup88 In Complex With Nup98 C-Terminal Autoproteolytic Domain
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-06-15 Classification: TRANSPORT PROTEIN |
 |
Crystal Structure Of The N-Terminal Domain Of Nup358/Ranbp2 (Residues 145-673)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2022-06-15 Classification: TRANSFERASE |
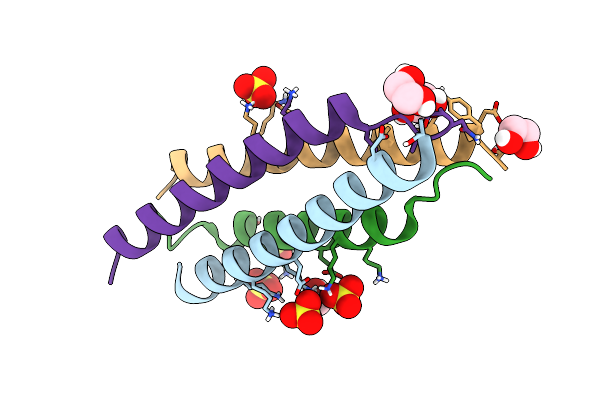 |
Crystal Structure Of The Tetramerization Element Of Nup358/Ranbp2 (Residues 805-832)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2022-06-15 Classification: TRANSPORT PROTEIN Ligands: EDO, SO4 |
 |
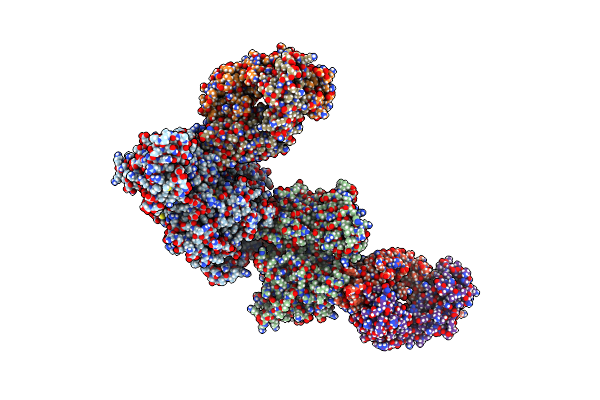
Crystal Structure Of The N-Terminal Domain Of Nup358/Ranbp2 (Residues 1-752) In Complex With Fab Fragment
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.95 Å Release Date: 2022-06-15 Classification: TRANSFERASE/Immune System |
 |
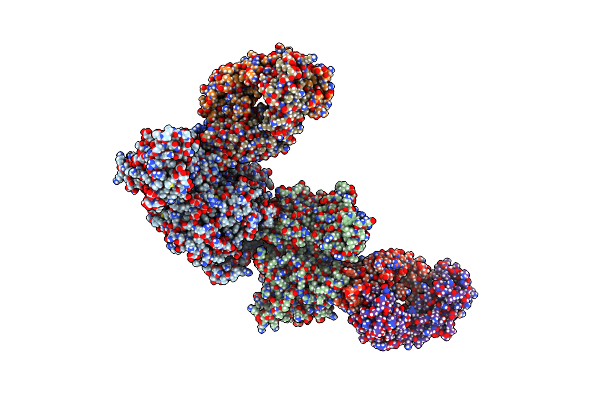
Crystal Structure Of The N-Terminal Domain Of Nup358/Ranbp2 (Residues 1-752) T585M Mutant In Complex With Fab Fragment
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:4.70 Å Release Date: 2022-06-15 Classification: TRANSPORT PROTEIN/Immune System |
 |
Crystal Structure Of The N-Terminal Domain Of Nup358/Ranbp2 (Residues 1-752) T653I Mutant In Complex With Fab Fragment
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:6.70 Å Release Date: 2022-06-15 Classification: TRANSPORT PROTEIN/Immune System |
 |
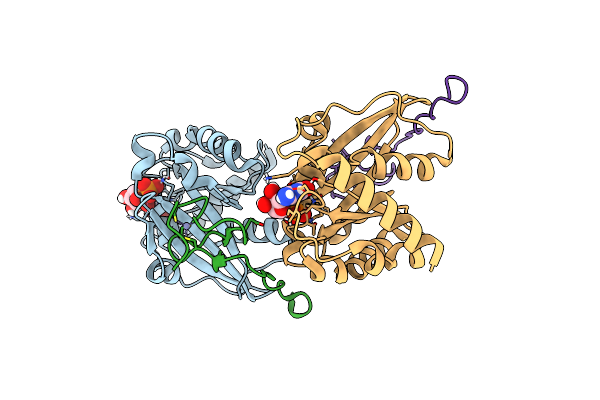
Crystal Structure Of The N-Terminal Domain Of Nup358/Ranbp2 (Residues 1-752) I656V Mutant In Complex With Fab Fragment
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:6.73 Å Release Date: 2022-06-15 Classification: TRANSFERASE/Immune System |
 |
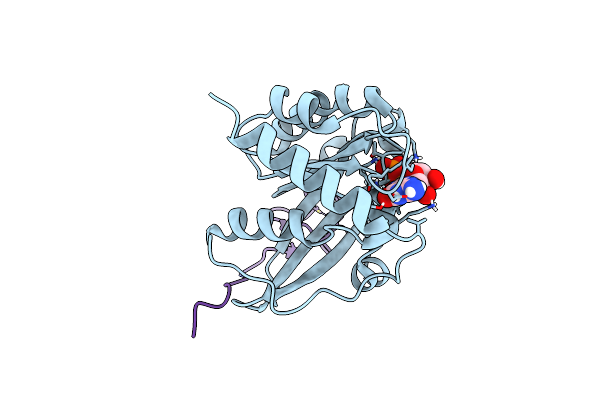
Crystal Structure Of The Znf2 Of Nucleoporin Nup358/Ranbp2 In Complex With Ran-Gdp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-06-15 Classification: TRANSPORT PROTEIN Ligands: GDP, MG, ZN |
 |
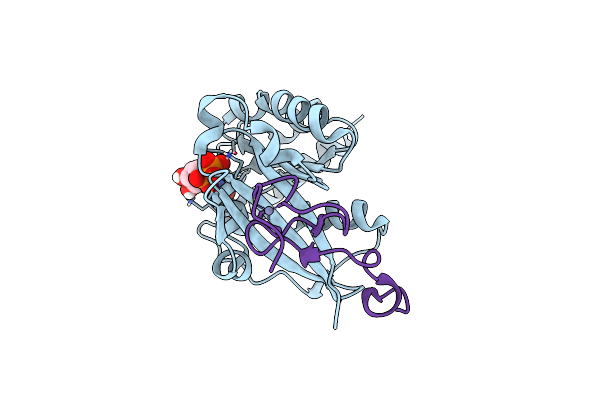
Crystal Structure Of The Znf2 Of Nucleoporin Nup358/Ranbp2 In Complex With Ran-Gdp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-06-15 Classification: TRANSPORT PROTEIN Ligands: GDP, MG, ZN |
 |
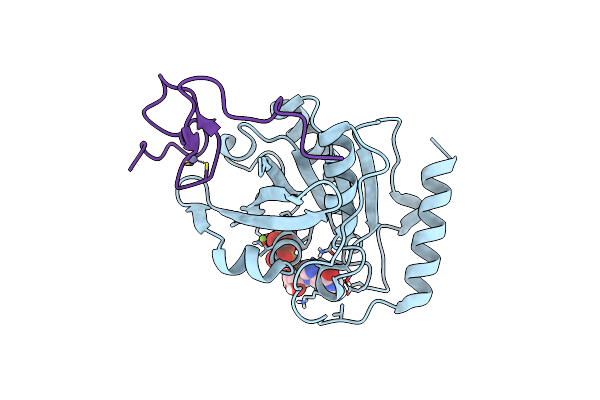
Crystal Structure Of The Znf3 Of Nucleoporin Nup358/Ranbp2 In Complex With Ran-Gdp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-06-15 Classification: TRANSPORT PROTEIN Ligands: GDP, MG, ZN |
 |
Crystal Structure Of The Znf4 Of Nucleoporin Nup358/Ranbp2 In Complex With Ran-Gdp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-06-15 Classification: TRANSPORT PROTEIN Ligands: GDP, MG, ZN |

