Search Count: 23
 |
Organism: Ruminiclostridium thermocellum atcc 27405
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-08-01 Classification: SUGAR BINDING PROTEIN Ligands: SO4, EDO |
 |
Organism: Ruminiclostridium thermocellum dsm 1313
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2018-01-31 Classification: HYDROLASE Ligands: 33O |
 |
Crystal Structure Of The Cohscac2-Xdoccipa Type Ii Complex From Clostridium Thermocellum
Organism: Ruminiclostridium thermocellum ad2, Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2017-04-05 Classification: CARBOHYDRATE BINDING PROTEIN Ligands: CA |
 |
Organism: Ruminiclostridium thermocellum dsm 1313
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-04-05 Classification: HYDROLASE Ligands: ZN, UNX, ADN |
 |
Organism: Ruminiclostridium thermocellum 27405, Clostridium thermocellum 27405
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2017-03-29 Classification: STRUCTURAL PROTEIN Ligands: CA |
 |
Determining The Specificities Of The Catalytic Site From The Very High Resolution Structure Of The Thermostable Glucuronoxylan Endo-Beta-1, 4-Xylanase, Ctxyn30A, From Clostridium Thermocellum With A Xylotetraose Bound
Organism: Ruminiclostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2016-10-19 Classification: HYDROLASE Ligands: PO4, PEG |
 |
The Mechanism By Which Arabinoxylanases Can Recognise Highly Decorated Xylans
Organism: Ruminiclostridium thermocellum jw20
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2016-08-31 Classification: HYDROLASE Ligands: SO4, CA, ARA |
 |
The Mechanism By Which Arabinoxylanases Can Recognise Highly Decorated Xylans
Organism: Ruminiclostridium thermocellum jw20
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-08-31 Classification: HYDROLASE Ligands: XYP, CA, 144 |
 |
The Mechanism By Which Arabinoxylanases Can Recognise Highly Decorated Xylans
Organism: Ruminiclostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2016-08-31 Classification: HYDROLASE Ligands: ARB, CA |
 |
Structure Of Fae Solved By Sad From Data Collected By Direct Data Collection (Ddc) Using The Esrf Robodiff Goniometer
Organism: Ruminiclostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2016-03-16 Classification: HYDROLASE Ligands: CD |
 |
Organism: Ruminiclostridium thermocellum atcc 27405
Method: X-RAY DIFFRACTION Resolution:3.61 Å Release Date: 2015-07-22 Classification: TRANSPORT PROTEIN/HYDROLASE |
 |
Crystal Structure Of The Peptidase-Containing Abc Transporter Pcat1 E648Q Mutant Complexed With Atpgs In An Occluded Conformation
Organism: Ruminiclostridium thermocellum atcc 27405
Method: X-RAY DIFFRACTION Resolution:5.52 Å Release Date: 2015-07-22 Classification: TRANSPORT PROTEIN/HYDROLASE Ligands: AGS |
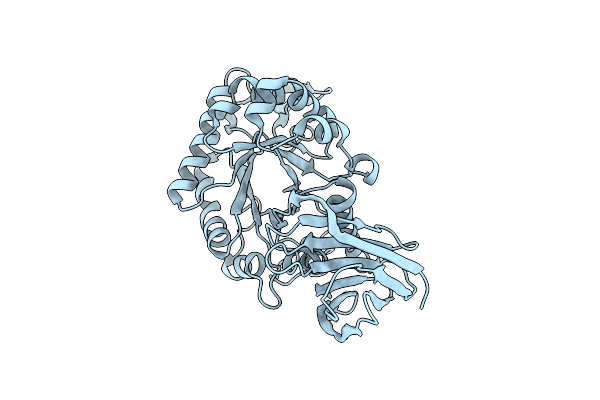 |
X-Ray Structure Of Glucuronoxylan-Xylanohydrolase (Xyn30A) From Clostridium Thermocellum At 1.77 A Resolution
Organism: Ruminiclostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2015-06-24 Classification: HYDROLASE |
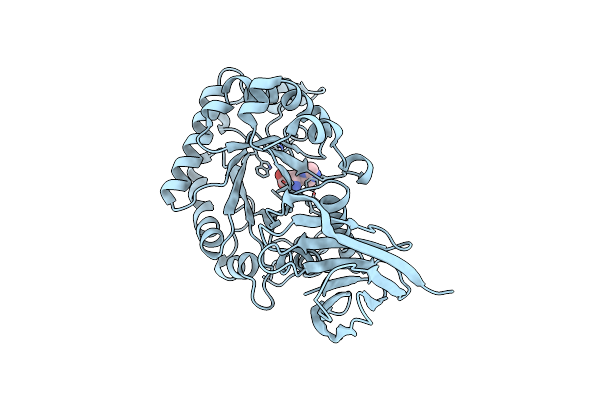 |
X-Ray Structure Of Glucuronoxylan-Xylanohydrolase (Xyn30A) From Clostridium Thermocellum
Organism: Ruminiclostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2015-06-24 Classification: HYDROLASE Ligands: HIS |
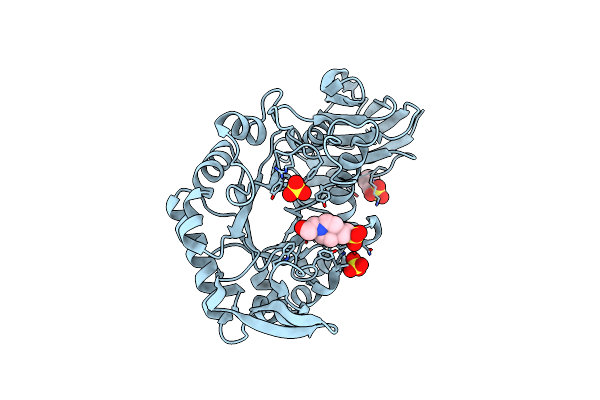 |
X-Ray Structure Of Glucuronoxylan-Xylanohydrolase (Xyn30A) From Clostridium Thermocellum At 1.68 A Resolution
Organism: Ruminiclostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2015-06-24 Classification: HYDROLASE Ligands: SO4, EPE |
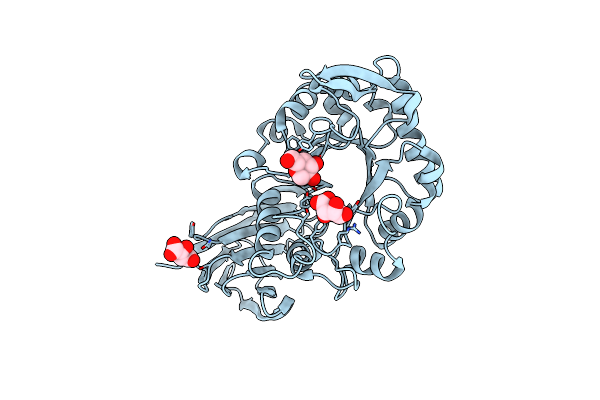 |
X-Ray Structure Of Glucuronoxylan-Xylanohydrolase (Xyn30A) From Clostridium Thermocellum At 1.30 A Resolution
Organism: Ruminiclostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2015-06-24 Classification: HYDROLASE Ligands: TAR, TLA, GLC |
 |
X-Ray Structure Of Glucuronoxylan-Xylanohydrolase (Xyn30A) From Clostridium Thermocellum At 1.25 A Resolution
Organism: Ruminiclostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2015-06-24 Classification: HYDROLASE Ligands: GOL, TRS, THR |
 |
X-Ray Structure Of Glucuronoxylan-Xylanohydrolase (Xyn30A) From Clostridium Thermocellum At 1.28 A Resolution
Organism: Ruminiclostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2015-06-24 Classification: HYDROLASE Ligands: GOL, TRS, PEG, PGO |
 |
Crystal Structure Of A Hypothetical Protein Cthe_0052 From Ruminiclostridium Thermocellum Atcc 27405
Organism: Ruminiclostridium thermocellum atcc 27405
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2015-01-28 Classification: UNKNOWN FUNCTION Ligands: TRS, PG4, FLC |
 |
Organism: Ruminiclostridium thermocellum
Method: SOLUTION NMR Release Date: 2014-10-15 Classification: HYDROLASE Ligands: CA |

