Search Count: 14
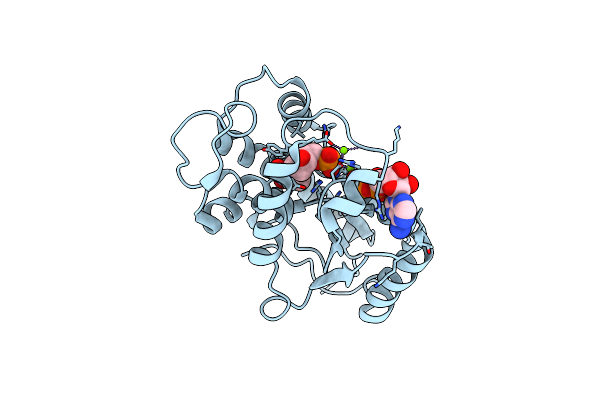 |
Structure Of Thymidylate Kinase From Candida Albicans Reveals Origin Of Broad Substrate Specificity And A Novel Structural Element.
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2017-08-09 Classification: TRANSFERASE Ligands: TMP, ADP, MG |
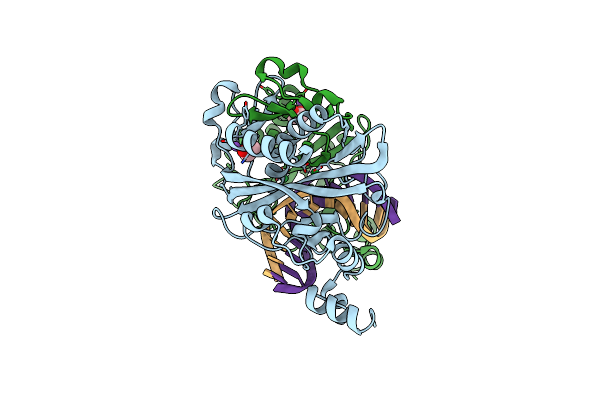 |
Crystal Structure Of The Ternary Ecorv-Dna-Lu Complex With Uncleaved Dna Substrate. Lanthanide Binding To Ecorv-Dna Complex Inhibits Cleavage.
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2016-11-09 Classification: HYDROLASE/DNA Ligands: LU, NA, EDO |
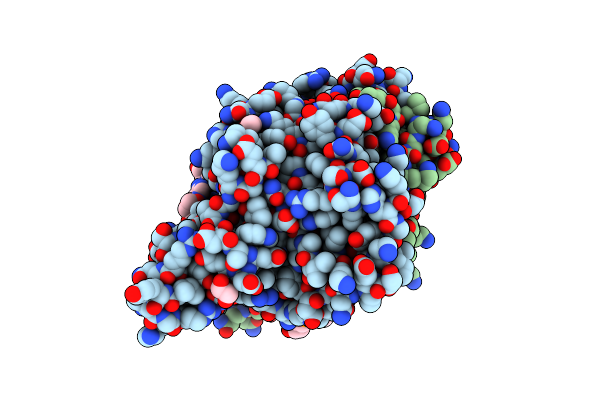 |
Crystal Structure Of The Ternary Ecorv-Dna-Lu Complex With Cleaved Dna Substrate.
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-11-09 Classification: HYDROLASE/DNA Ligands: EDO, NA, LU |
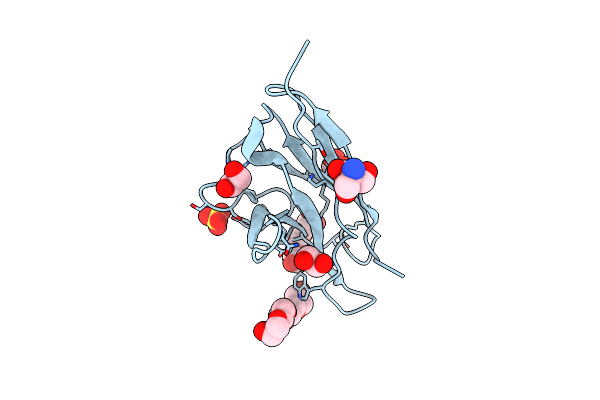 |
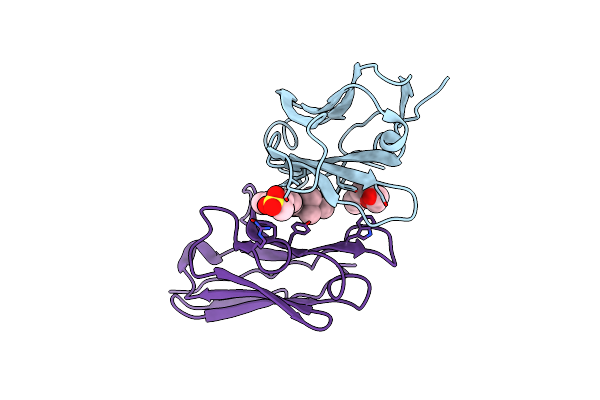
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2012-03-21 Classification: DYE-BINDING PROTEIN Ligands: 1PE, PE3, EDO, SO4, TRS |
 |
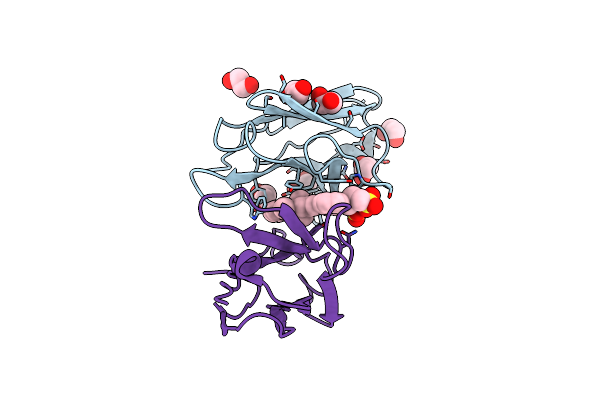
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-03-21 Classification: DYE-BINDING PROTEIN Ligands: CL, PE3, DIW |
 |
Fluorogen Activating Protein M8Vla4(S55P) In Complex With Dimethylindole Red
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2012-03-21 Classification: DYE-BINDING PROTEIN Ligands: DIW, EDO, SO4 |
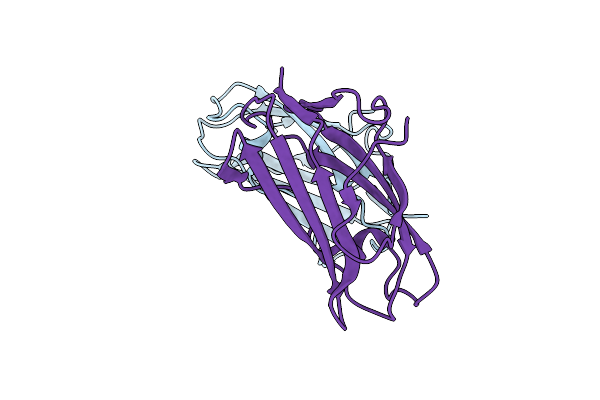 |
Organism: Dermatophagoides pteronyssinus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2002-05-15 Classification: ALLERGEN |
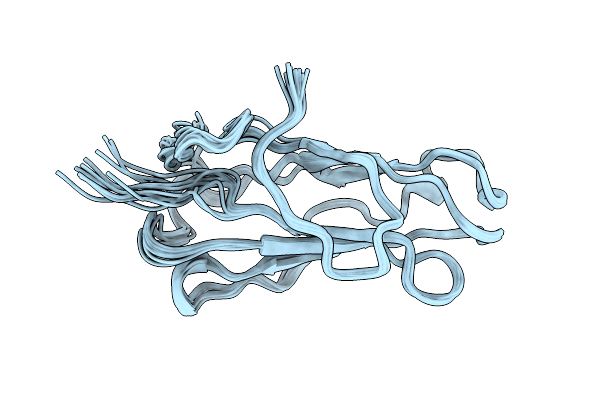 |
Tertiary Structure Of The Major House Dust Mite Allergen Der P 2, Nmr, 10 Structures
Organism: Dermatophagoides pteronyssinus
Method: SOLUTION NMR Release Date: 1998-10-14 Classification: ALLERGEN |
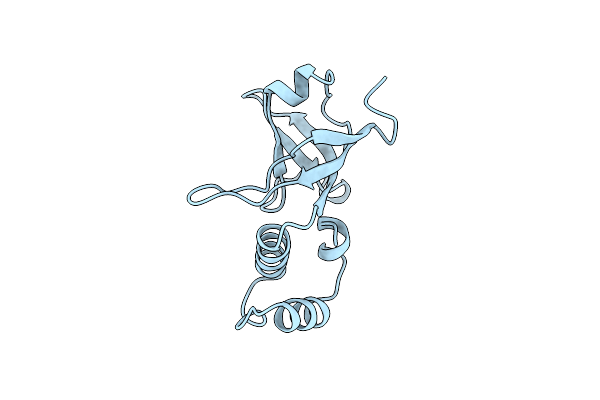 |
Crystal Structure Of The Rna-Binding Domain Of The Transcriptional Terminator Protein Rho
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 1998-06-17 Classification: TRANSCRIPTION TERMINATION |
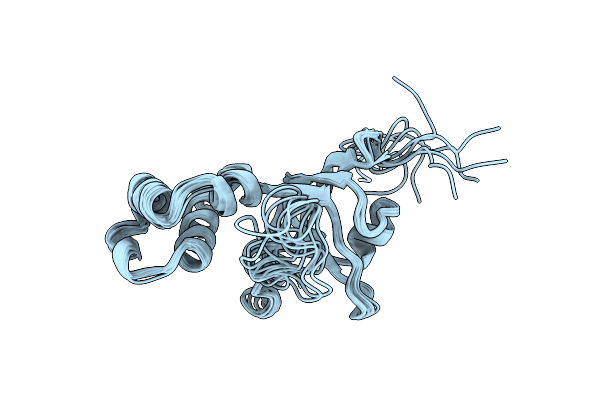 |
The Nmr Structure Of The Rna Binding Domain Of E.Coli Rho Factor Suggests Possible Rna-Protein Interactions, 10 Structures
Organism: Escherichia coli bl21(de3)
Method: SOLUTION NMR Release Date: 1998-05-27 Classification: TRANSCRIPTION TERMINATION |
 |
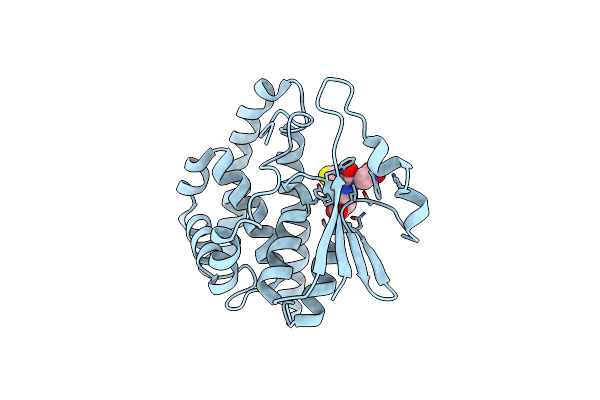 |
Crystal Structure Of Human Class Mu Glutathione Transferase Gstm2-2: Effects Of Lattice Packing On Conformational Heterogeneity
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 1994-01-31 Classification: TRANSFERASE(GLUTATHIONE) Ligands: GDN |
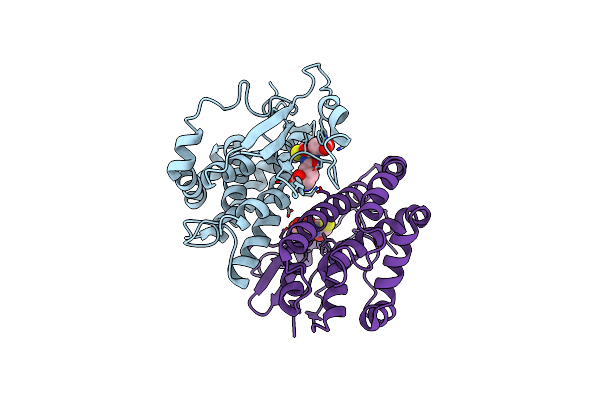 |
Crystal Structure Of Human Class Mu Glutathione Transferase Gstm2-2: Effects Of Lattice Packing On Conformational Heterogeneity
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 1994-01-31 Classification: TRANSFERASE(GLUTATHIONE) Ligands: GDN |
 |
Crystal Structure Of Human Class Mu Glutathione Transferase Gstm2-2: Effects Of Lattice Packing On Conformational Heterogeneity
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 1994-01-31 Classification: TRANSFERASE(GLUTATHIONE) Ligands: GDN |

