Search Count: 19
 |
Organism: Myroides odoratus
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: VIRAL PROTEIN |
 |
Organism: Bacillus amyloliquefaciens
Method: X-RAY DIFFRACTION Release Date: 2025-04-16 Classification: ANTIVIRAL PROTEIN Ligands: Y65, SO4 |
 |
Structure Of Myroides Odoratus Prophage Anti-Thoeris 2 (Modtad2) In Complex With Hisadpr
Organism: Myroides odoratus
Method: X-RAY DIFFRACTION Release Date: 2025-04-16 Classification: VIRAL PROTEIN Ligands: Y65 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2024-12-18 Classification: LYASE Ligands: ZN, PPI, TRS, DMS, EDO, VV8, ACY |
 |
Three-Dimensional Structure Of Human Carbonic Anhydrase Ix In Complex With A Covalent Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-12-18 Classification: LYASE Ligands: ZN, A1IDV |
 |
Organism: Candidatus cloacimonas acidaminovorans, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: HYDROLASE |
 |
Organism: Candidatus cloacimonas acidaminovorans, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: HYDROLASE |
 |
Organism: Candidatus cloacimonas acidaminovorans, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: HYDROLASE |
 |
Three-Dimensional Structure Of Human Carbonic Anhydrase Ii In Complex With A Covalent Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-04-10 Classification: LYASE Ligands: ZN, VV8 |
 |
Organism: Bacillus cereus msx-d12
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2024-02-21 Classification: HYDROLASE |
 |
Organism: Bacillus cereus msx-d12
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: HYDROLASE Ligands: OJC, NAD |
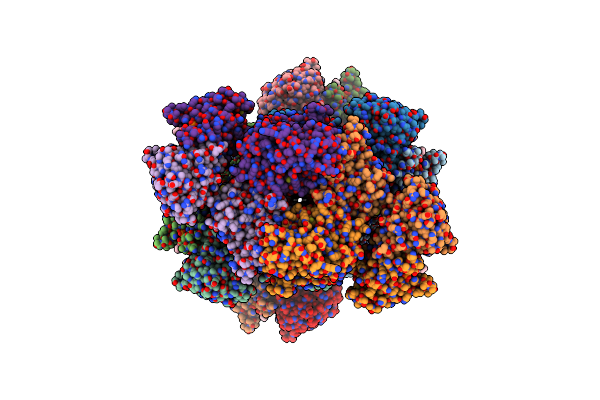 |
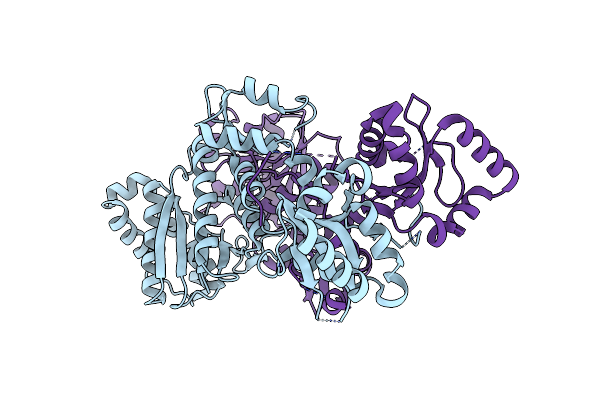
Organism: Bacillus cereus msx-d12
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: HYDROLASE Ligands: RK3, ENA |
 |
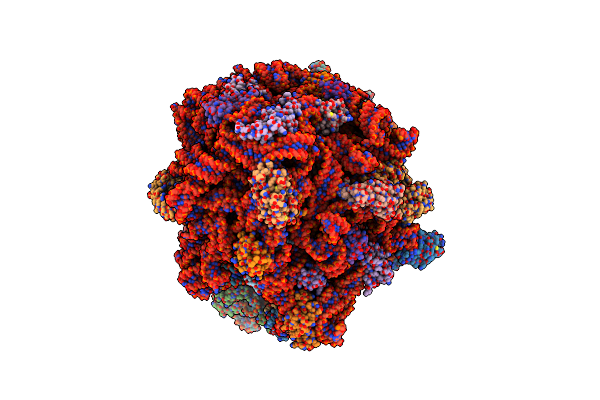
Organism: Allochromatium vinosum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-12-13 Classification: HYDROLASE |
 |
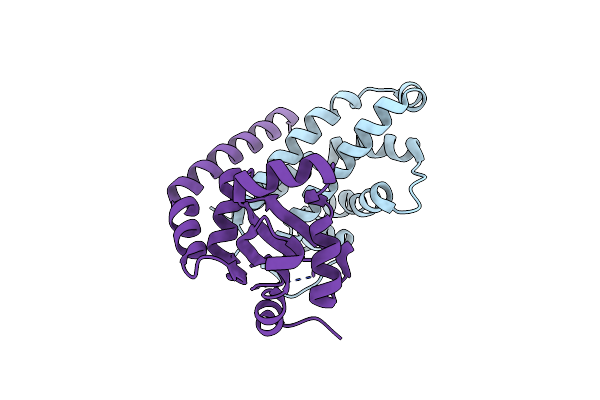
Organism: Allochromatium vinosum, Synthetic construct, Streptococcus thermophilus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-12-13 Classification: RIBOSOME Ligands: ZN, MG |
 |
Organism: Aphanizomenon flos-aquae 2012/km1/d3
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-12-30 Classification: TOXIN/ANTITOXIN |
 |
Organism: Aphanizomenon flos-aquae 2012/km1/d3
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-12-30 Classification: TOXIN Ligands: AMP, EDO |
 |
Organism: Aphanizomenon flos-aquae 2012/km1/d3
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-12-30 Classification: TOXIN Ligands: ACT |
 |
Crystal Structure Of Mono-Ampylated Hepn(R46E) Toxin In Complex With Mnt Antitoxin
Organism: Aphanizomenon flos-aquae 2012/km1/d3
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2020-12-30 Classification: TOXIN/ANTITOXIN Ligands: 2DA |
 |
Rebuilt And Re-Refined Pdb Entry 5Yep: Tri-Ampylated Shewanella Oneidensis Hepn Toxin In Complex With Mnt Antitoxin
Organism: Shewanella oneidensis (strain mr-1)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-12-30 Classification: TOXIN/ANTITOXIN Ligands: AMP |

