Search Count: 36
 |
Biological And Structural Analysis Of New Potent Integrase-Ledgf Allosteric Hiv-1 Inhibitors
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-06-07 Classification: VIRAL PROTEIN Ligands: RWR, MG, SO4 |
 |
Hiv-1 Integrase Catalytic Core Domain And C-Terminal Domain In Complex With Allosteric Integrase Inhibitor Bdm-2
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-06-07 Classification: VIRAL PROTEIN Ligands: RWR, EDO, CL, MG |
 |
Hiv-1 Integrase Catalytic Core Domain And C-Terminal Domain In Complex With Allosteric Integrase Inhibitor Mut871
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-06-07 Classification: VIRAL PROTEIN Ligands: EDO, CL, U5L, MG |
 |
Hiv-1 Integrase Catalytic Core Domain And C-Terminal Domain In Complex With Allosteric Integrase Inhibitor Mut872
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2023-06-07 Classification: VIRAL PROTEIN Ligands: W2Q, EDO, MG, PEG, CL |
 |
Hiv-1 Integrase Catalytic Core Domain And C-Terminal Domain In Complex With Allosteric Integrase Inhibitor Mut884
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2023-06-07 Classification: VIRAL PROTEIN Ligands: CL, EDO, U60, MG, PGE |
 |
Hiv-1 Integrase Catalytic Core Domain And C-Terminal Domain In Complex With Allosteric Integrase Inhibitor Mut916
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2023-06-07 Classification: VIRAL PROTEIN Ligands: EDO, PEG, U5S, MG, CL |
 |
Crystal Structure Of Lugdulysin, A Staphylococcus Lugdunensis M30 Zinc Metallopeptidase
Organism: Staphylococcus lugdunensis
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2022-07-13 Classification: HYDROLASE Ligands: ZN, CA, CAC, ACT |
 |
Organism: Staphylococcus lugdunensis
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2022-07-13 Classification: HYDROLASE Ligands: CA, ZN |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-07-22 Classification: VIRAL PROTEIN Ligands: NI |
 |
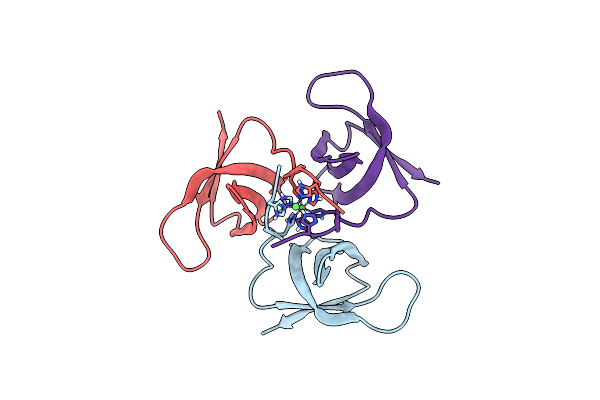
Crystal Structure Of The C-Terminal Domain Of The Hiv-1 Integrase (Subtype A2)
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-07-22 Classification: VIRAL PROTEIN Ligands: NI |
 |
Crystal Structure Of The C-Terminal Domain Of The Hiv-1 Integrase (Subtype A2, Mutant N254K, K340Q)
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-07-22 Classification: VIRAL PROTEIN Ligands: NI |
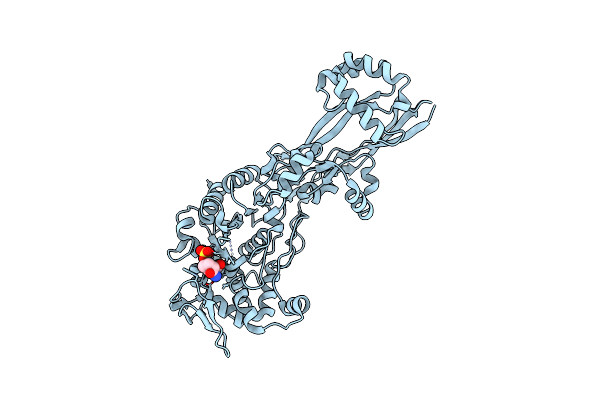 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2019-05-22 Classification: HYDROLASE/ANTIBIOTIC Ligands: NXL |
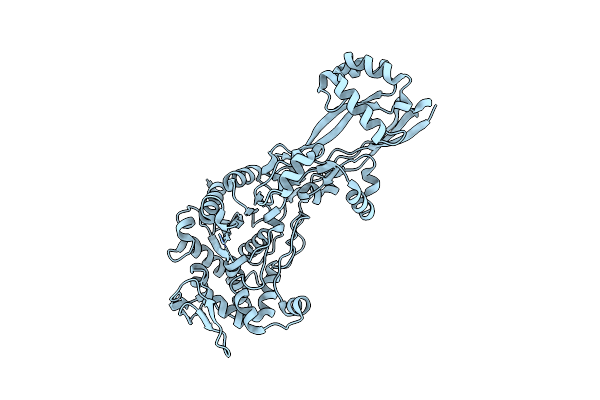 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-05-22 Classification: HYDROLASE/ANTIBIOTIC |
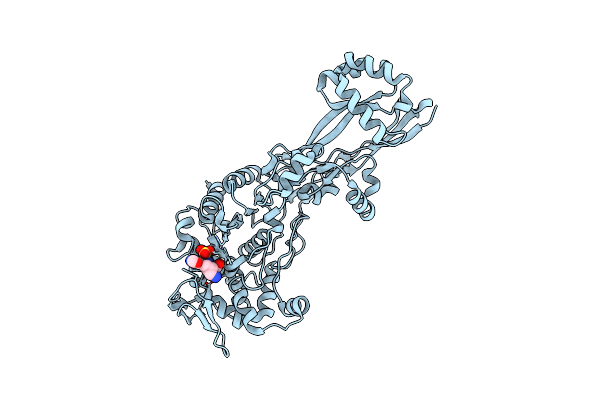 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-05-22 Classification: HYDROLASE/ANTIBIOTIC Ligands: ET5 |
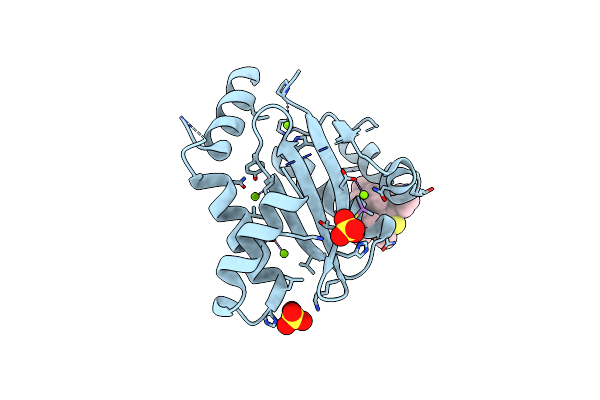 |
Dissociation Of Biochemical And Antiretroviral Activities Of Integrase-Ledgf Allosteric Inhibitors Revealed By Resistance Of A125 Polymorphic Hiv-1
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-03-07 Classification: VIRAL PROTEIN Ligands: MG, 9VN, SO4 |
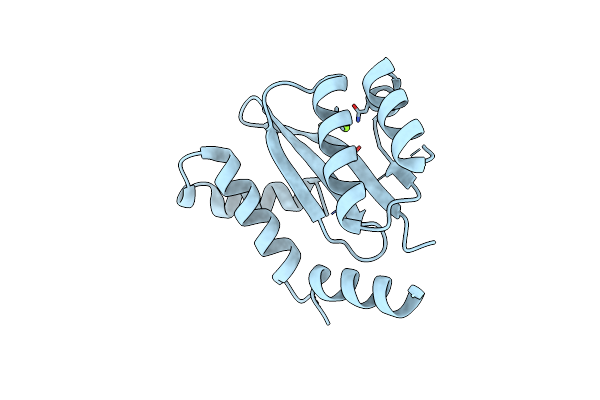 |
Dissociation Of Biochemical And Antiretroviral Activities Of Integrase-Ledgf Allosteric Inhibitors Revealed By Resistance Of A125 Polymorphic Hiv-1
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-03-07 Classification: VIRAL PROTEIN Ligands: MG |
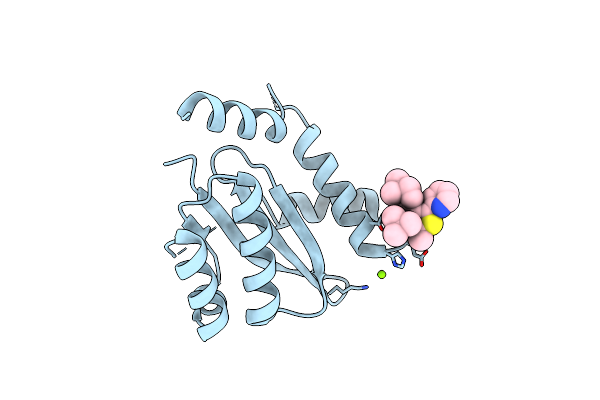 |
Dissociation Of Biochemical And Antiretroviral Activities Of Integrase-Ledgf Allosteric Inhibitors Revealed By Resistance Of A125 Polymorphic Hiv-1
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-03-07 Classification: VIRAL PROTEIN Ligands: 9VK, MG |
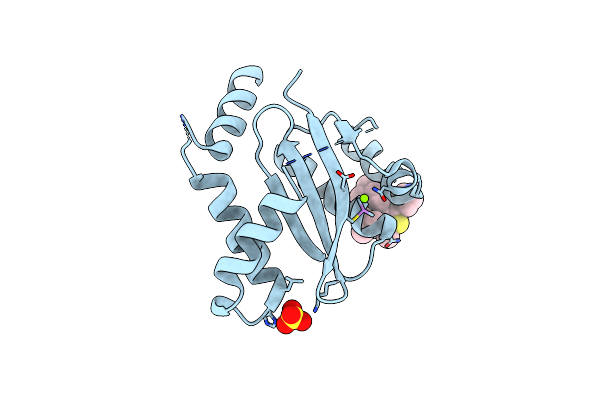 |
Dissociation Of Biochemical And Antiretroviral Activities Of Integrase-Ledgf Allosteric Inhibitors Revealed By Resistance Of A125 Polymorphic Hiv-1
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2018-03-07 Classification: VIRAL PROTEIN Ligands: MG, 9VN, SO4 |
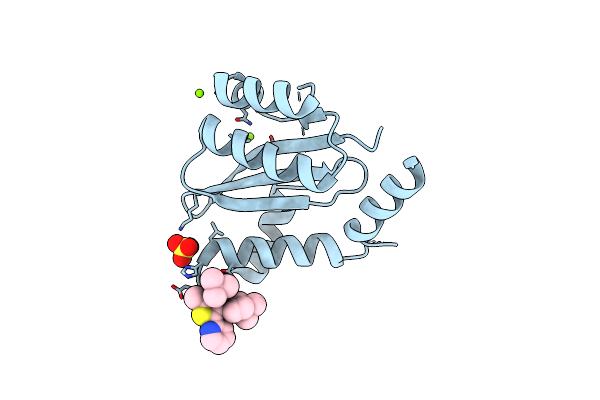 |
Dissociation Of Biochemical And Antiretroviral Activities Of Integrase-Ledgf Allosteric Inhibitors Revealed By Resistance Of A125 Polymorphic Hiv-1
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-03-07 Classification: VIRAL PROTEIN Ligands: MG, 9VK, SO4 |
 |
Glycation Restrains Allosteric Transition In Hemoglobin: The Molecular Basis Of Oxidative Stress Under Hyperglycemic Conditions In Diabetes
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-02-01 Classification: OXYGEN TRANSPORT Ligands: HEM, OXY, FRU, GLC |

