Search Count: 56
 |
Structure Of The Reconstructed Ancestor Of Phenolic Acid Decarboxylase Ancpad31
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-09-27 Classification: LYASE |
 |
Structure Of The Reconstructed Ancestor Of Phenolic Acid Decarboxylase Ancpad55
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-08-02 Classification: LYASE Ligands: SO4 |
 |
Structure Of The Reconstructed Ancestor Of Phenolic Acid Decarboxylase Ancpad55
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-07-19 Classification: LYASE Ligands: SO4 |
 |
Structure Of The Reconstructed Ancestor Of Phenolic Acid Decarboxylase Ancpad134
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2023-07-05 Classification: LYASE |
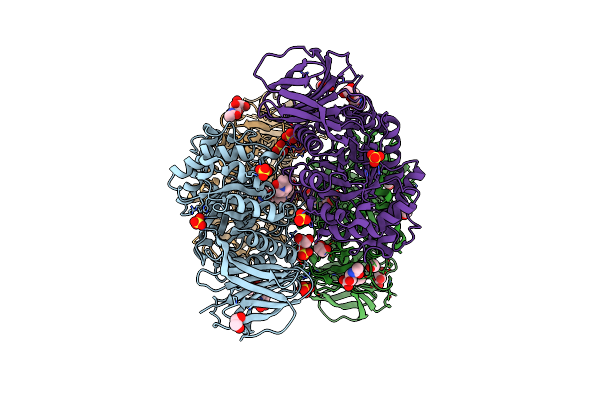 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-12-23 Classification: HYDROLASE Ligands: SO4, TRS, EDO, NAG, GOL, M7H, CL |
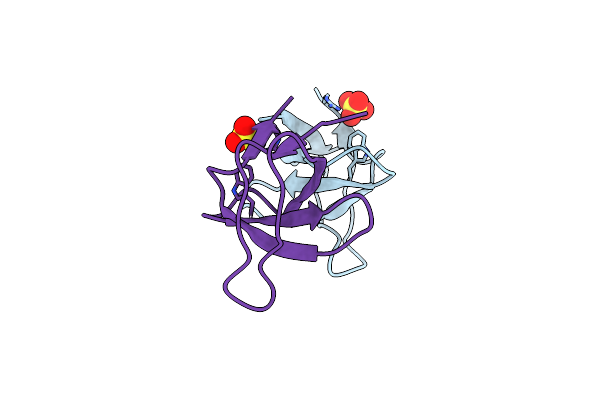 |
Crystal Structure Of The Monomeric Human Nck Sh3.1 Domain, Triclinic, 1.08A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2020-02-12 Classification: SIGNALING PROTEIN Ligands: SO4 |
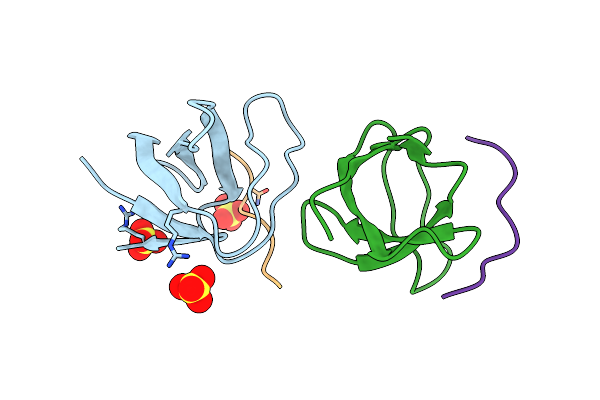 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.04 Å Release Date: 2020-02-12 Classification: SIGNALING PROTEIN Ligands: SO4 |
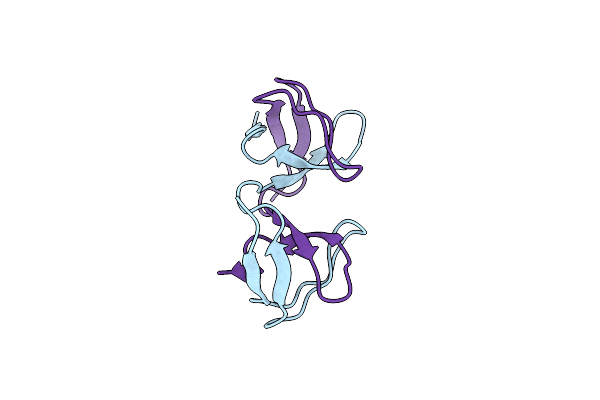 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2020-02-12 Classification: SIGNALING PROTEIN |
 |
Crystal Structure Of Swapped Human Nck Sh3.1 Domain, 1.05A, Orthorhombic Form I
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2020-02-12 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.11 Å Release Date: 2020-02-12 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2020-02-12 Classification: SIGNALING PROTEIN |
 |
Crystal Structure Of Swapped Human Nck Sh3.1 Domain, 1.3A, Orthorhombic Form Iii
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2020-02-12 Classification: SIGNALING PROTEIN |
 |
Crystal Structure Of Symmetric Swapped Human Nck Sh3.1 Domain, 0.93A, Orthorhombic Form Iv
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:0.93 Å Release Date: 2020-02-12 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2020-02-12 Classification: SIGNALING PROTEIN |
 |
Crystal Structure Of Domain-Swapped C-Terminal Domain Of Human Doublecortin
Organism: Homo sapiens, Unidentified
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2018-04-04 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2017-09-27 Classification: HYDROLASE Ligands: CL |
 |
Crystal Structure Of Bace-1 In Complex With Active Site Inhibitor Grl-8234 And Exosite Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2017-09-27 Classification: HYDROLASE Ligands: BSD |
 |
Crystal Structure Of Bace-1 In Complex With Active Site And Exosite Binding Peptide Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2017-09-27 Classification: HYDROLASE Ligands: EDO |
 |
Crystal Structure Of Fab Of Parasite Invasion Inhibitory Antibody C1 - Hexagonal Form
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2017-01-25 Classification: IMMUNE SYSTEM Ligands: CL |
 |
Crystal Structure Of Fab Of Parasite Invasion Inhibitory Antibody C1 - Orthorhombic Form
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-01-25 Classification: IMMUNE SYSTEM |

