Search Count: 13
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-09-20 Classification: MEMBRANE PROTEIN |
 |
Structural Coordination Of Polymerization And Crosslinking By A Peptidoglycan Synthase Complex
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2020-03-18 Classification: MEMBRANE PROTEIN |
 |
Structural Coordination Of Polymerization And Crosslinking By A Peptidoglycan Synthase Complex
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2020-03-18 Classification: MEMBRANE PROTEIN Ligands: AIX |
 |
Organism: bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2018-10-10 Classification: STRUCTURAL PROTEIN |
 |
Tandem Germn Domains Of The Sporulation Protein Germ From Bacillus Subtilis
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-10-10 Classification: STRUCTURAL PROTEIN Ligands: ACT, EDO |
 |
Crystal Structure Of Thermus Thermophilus Rod Shape Determining Protein Roda (Q5Six3_Thet8)
Organism: Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579)
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2018-03-28 Classification: TRANSFERASE Ligands: OLC, CL |
 |
Crystal Structure Of Thermus Thermophilus Rod Shape Determining Protein Roda D255A Mutant (Q5Six3_Thet8)
Organism: Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579)
Method: X-RAY DIFFRACTION Resolution:3.19 Å Release Date: 2018-03-28 Classification: TRANSFERASE Ligands: CL |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-12-04 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2013-12-04 Classification: HYDROLASE/PEPTIDE |
 |
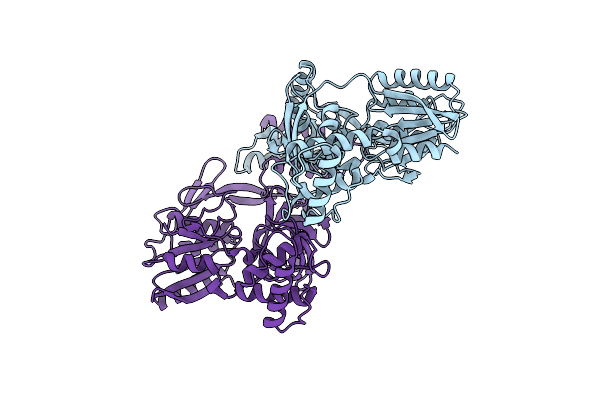
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-12-04 Classification: HYDROLASE |
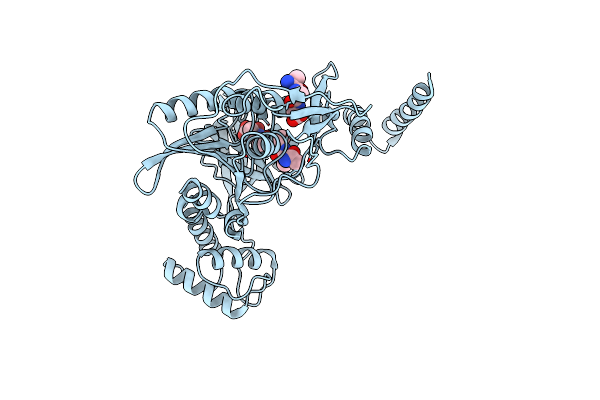 |
Crystal Structure Of The Ctpb R168A Mutant Present In An Active Conformation
Organism: Bacillus subtilis subsp. subtilis str. 168, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2013-12-04 Classification: HYDROLASE |
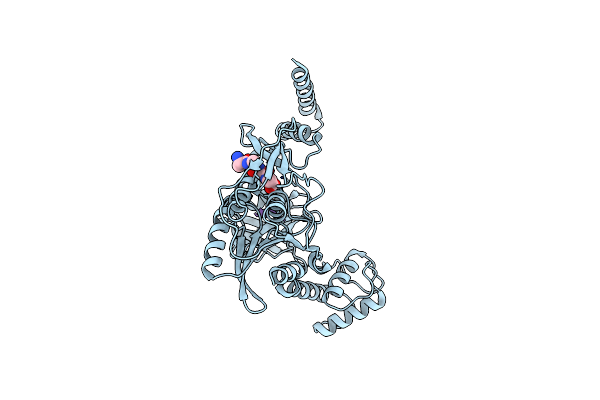 |
Crystal Structure Of Ctpb(S309A) In Complex With A Peptide Having A Val-Pro-Ala C-Terminus
Organism: Bacillus subtilis subsp. subtilis str. 168, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-12-04 Classification: HYDROLASE/PEPTIDE |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-12-04 Classification: HYDROLASE |

