Search Count: 17
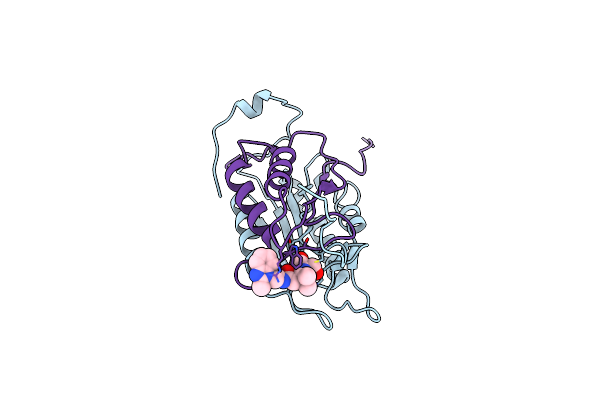 |
Succinic Acid Amides As P2-P3 Replacements For Inhibitors Of Interleukin-1Beta Converting Enzyme (Ice Or Caspase 1)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2011-08-17 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 3NS |
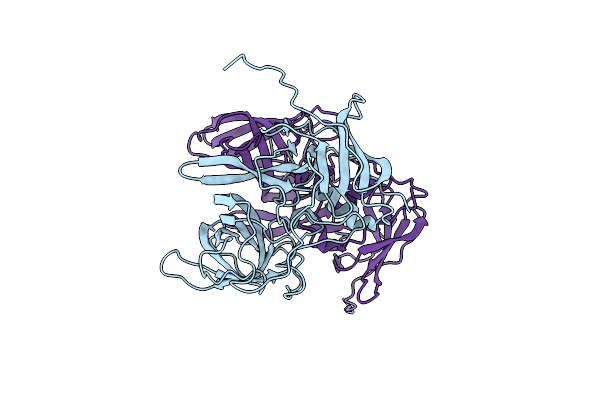 |
Organism: Murine norovirus 1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-04-21 Classification: VIRAL PROTEIN |
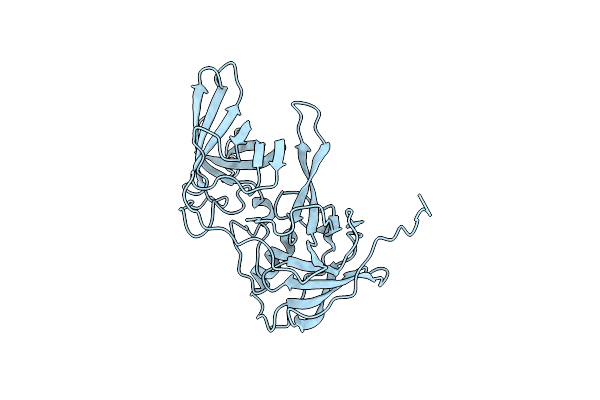 |
X-Ray Structure Of The Murine Norovirus (Mnv)-1 Capsid Protein Protruding (P) Domain
Organism: Murine norovirus 1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-04-21 Classification: VIRAL PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2009-09-01 Classification: HYDROLASE Ligands: MG, CL |
 |
Crystal Structure Of Yrbi Lacking The Last 8 Residues, In Complex With Kdo And Inorganic Phosphate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2009-09-01 Classification: HYDROLASE Ligands: MG, PO4, KDO |
 |
Organism: Escherichia coli o6
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2008-09-23 Classification: HYDROLASE Ligands: MG, CL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2008-09-23 Classification: HYDROLASE Ligands: CL |
 |
Crystal Structure Of Yrbi Phosphatase From Escherichia Coli In A Complex With Ca
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2008-09-23 Classification: HYDROLASE Ligands: CA, CL |
 |
Crystal Structure Of Yrbi Phosphatase From Escherichia Coli In Complex With A Phosphate And A Calcium Ion
Organism: Escherichia coli o6
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-09-23 Classification: HYDROLASE Ligands: CA, PO4 |
 |
Crystal Structure Of Human Branched Chain Amino Acid Transaminase In A Complex With An Inhibitor, C16H10N2O4F3Scl, And Pyridoxal 5' Phosphate.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2006-06-27 Classification: TRANSFERASE Ligands: CBC, PLP |
 |
Crystal Structure Of Thrombin In Complex With A Novel Bicyclic Lactam Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2002-02-27 Classification: HYDROLASE Ligands: BLI |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 1999-11-19 Classification: HYDROLASE Ligands: ZN, CA |
 |
Hairpin Loop Containing Domain Of Hepatocyte Growth Factor, Nmr, Minimized Average Structure
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 1998-06-24 Classification: HEPATOCYTE GROWTH FACTOR |
 |
Organism: Rous sarcoma virus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1997-07-23 Classification: V-SRC SH2 DOMAIN |
 |
Organism: Rous sarcoma virus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1997-07-07 Classification: TRANSFORMING PROTEIN Ligands: 1C5 |
 |
Crystal Structure Of Recombinant Bovine Interferon-Gamma At 3.0 Angstroms Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 1994-01-31 Classification: GLYCOPROTEIN |
 |
Crystal Structure Of Recombinant Rabbit Interferon-Gamma At 2.7-Angstroms Resolution
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1993-01-15 Classification: CYTOKINE |

