Search Count: 20
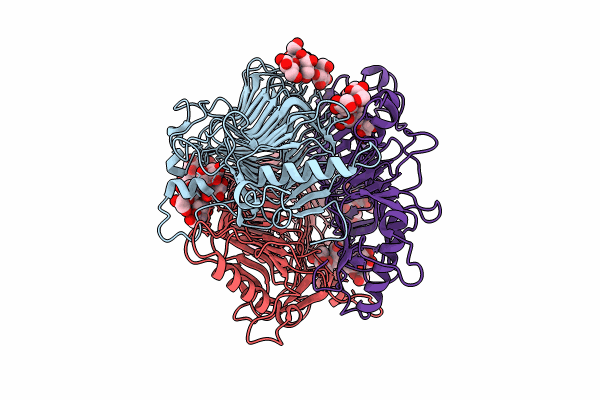 |
Crystal Structure Of Gh9L Inulin Fructotransferases (Iftase) In Compex With Fruetosyl Nystose (Gf4)
Organism: Paenarthrobacter aurescens
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2024-09-04 Classification: CARBOHYDRATE |
 |
Crystal Structure Of Gh9L Inulin Fructotransferases(Iftase)Incomplex With Nystose(F3)
Organism: Paenarthrobacter aurescens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-09-04 Classification: CARBOHYDRATE Ligands: FRU |
 |
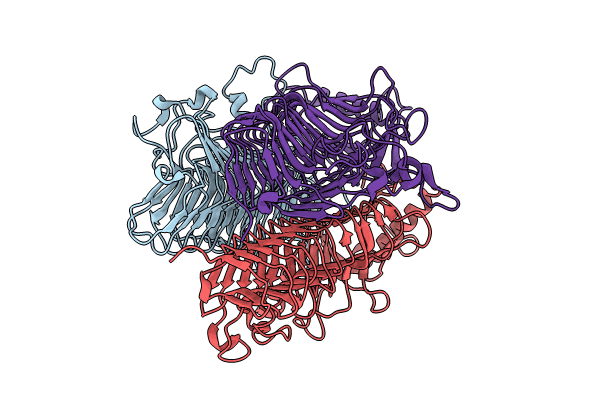
Crystal Structure Of Gh9L Inulinfructotransferases (Iftase) In Complex With Gf2
Organism: Paenarthrobacter aurescens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-09-04 Classification: CARBOHYDRATE |
 |
Organism: Paenarthrobacter aurescens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2024-09-04 Classification: CARBOHYDRATE |
 |
Organism: Legionella pneumophila subsp. pneumophila str. philadelphia 1, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2022-08-24 Classification: HYDROLASE Ligands: AYE |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-12-01 Classification: FLAVOPROTEIN Ligands: SF4, FMN, CL |
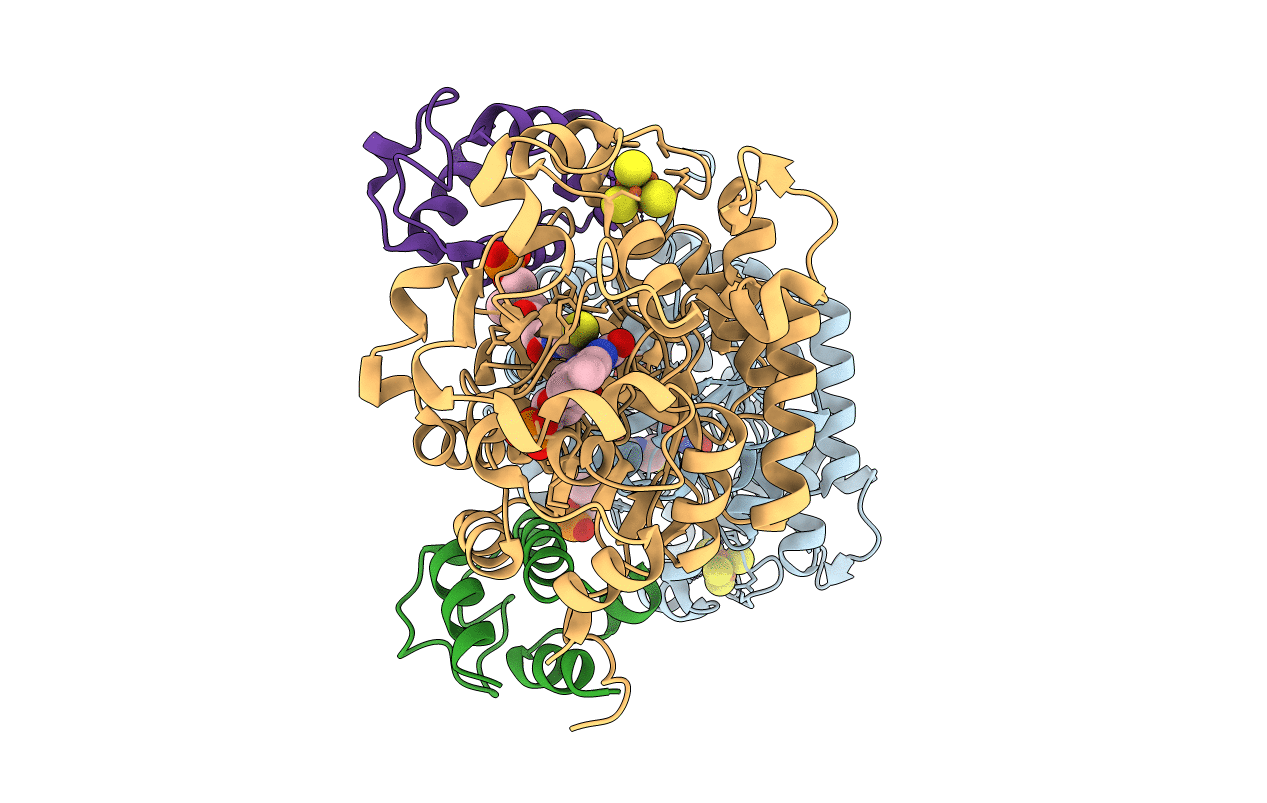 |
Crystal Structure Of Dehydrogenase/Isomerase Fabx From Helicobacter Pylori In Complex With Holo-Acp
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-12-01 Classification: FLAVOPROTEIN Ligands: FMN, SF4, PN7 |
 |
Crystal Structure Of Dehydrogenase/Isomerase Fabx From Helicobacter Pylori In Complex With Octanoyl-Acp
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2021-12-01 Classification: FLAVOPROTEIN Ligands: FMN, SF4, 66S |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-01-30 Classification: SIGNALING PROTEIN |
 |
Organism: Legionella longbeachae serogroup 1 (strain nsw150)
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2018-11-14 Classification: PEPTIDE BINDING PROTEIN Ligands: CL |
 |
Organism: Legionella longbeachae serogroup 1 (strain nsw150)
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2018-11-14 Classification: PEPTIDE BINDING PROTEIN Ligands: PTR |
 |
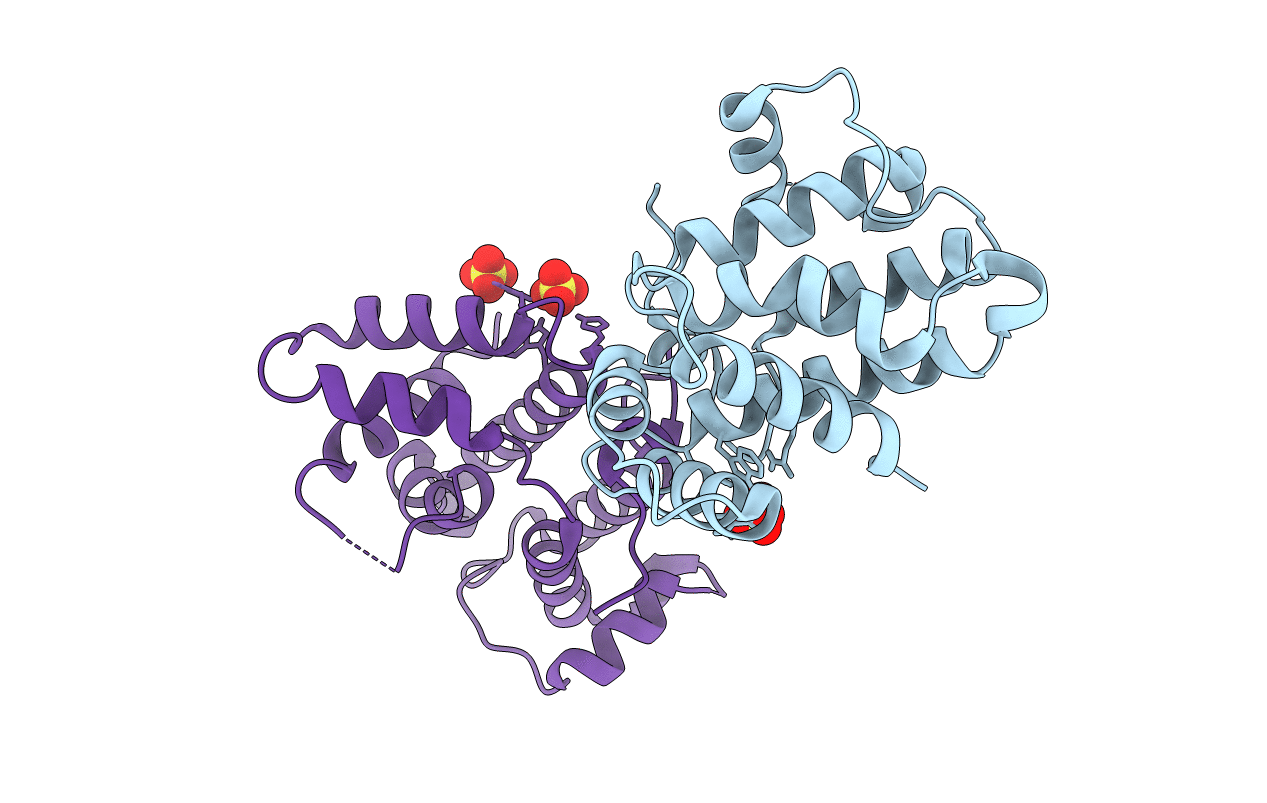
Legionella Longbeachae Lesh (Llo2327) Bound To The Human Interleukin-2 Receptor Beta Ptyr387 Peptide
Organism: Legionella longbeachae serogroup 1 (strain nsw150), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2018-11-14 Classification: PEPTIDE BINDING PROTEIN |
 |
Legionella Longbeachae Lesh (Llo2327) Bound To The Human Dnaj-A1 Ptyr381 Peptide
Organism: Legionella longbeachae serogroup 1 (strain nsw150), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2018-11-14 Classification: PEPTIDE BINDING PROTEIN |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2018-05-23 Classification: SIGNALING PROTEIN Ligands: SO4 |
 |
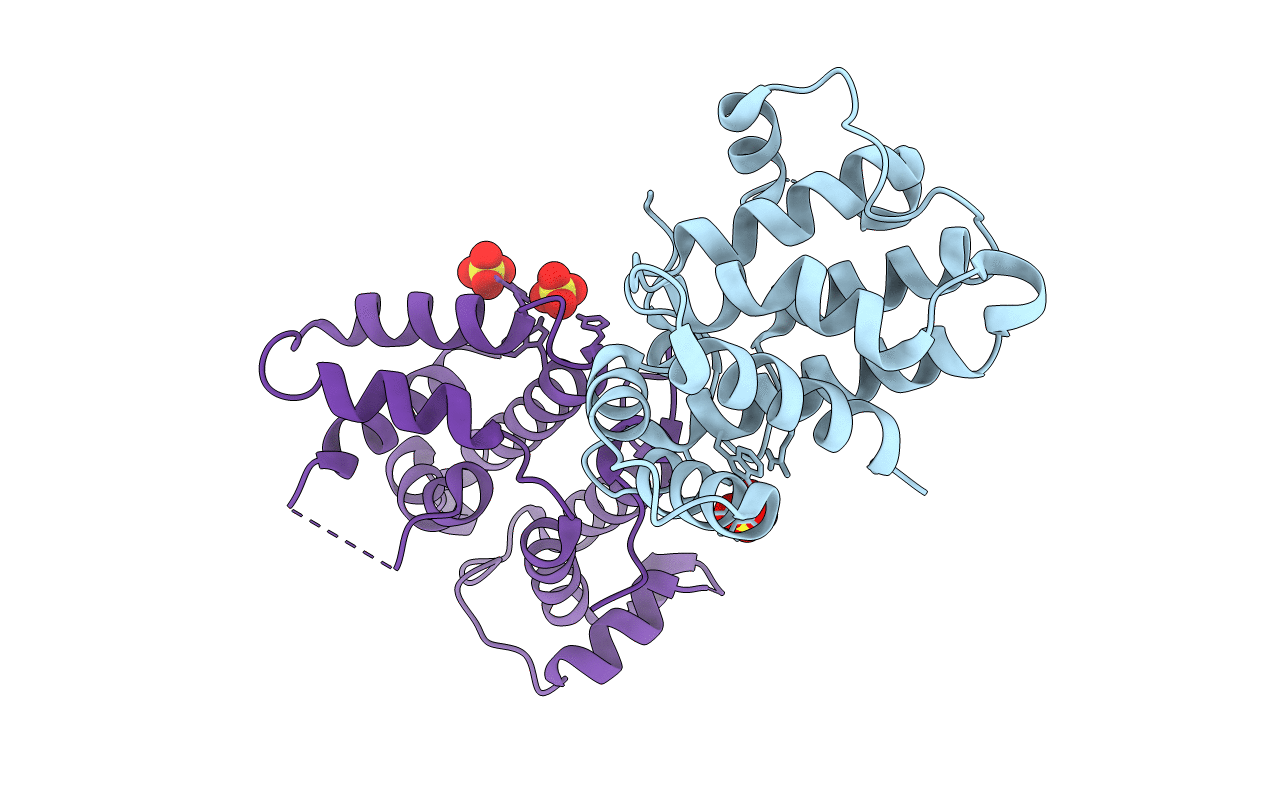
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2018-05-23 Classification: SIGNALING PROTEIN Ligands: SO4 |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-05-23 Classification: SIGNALING PROTEIN Ligands: SO4 |
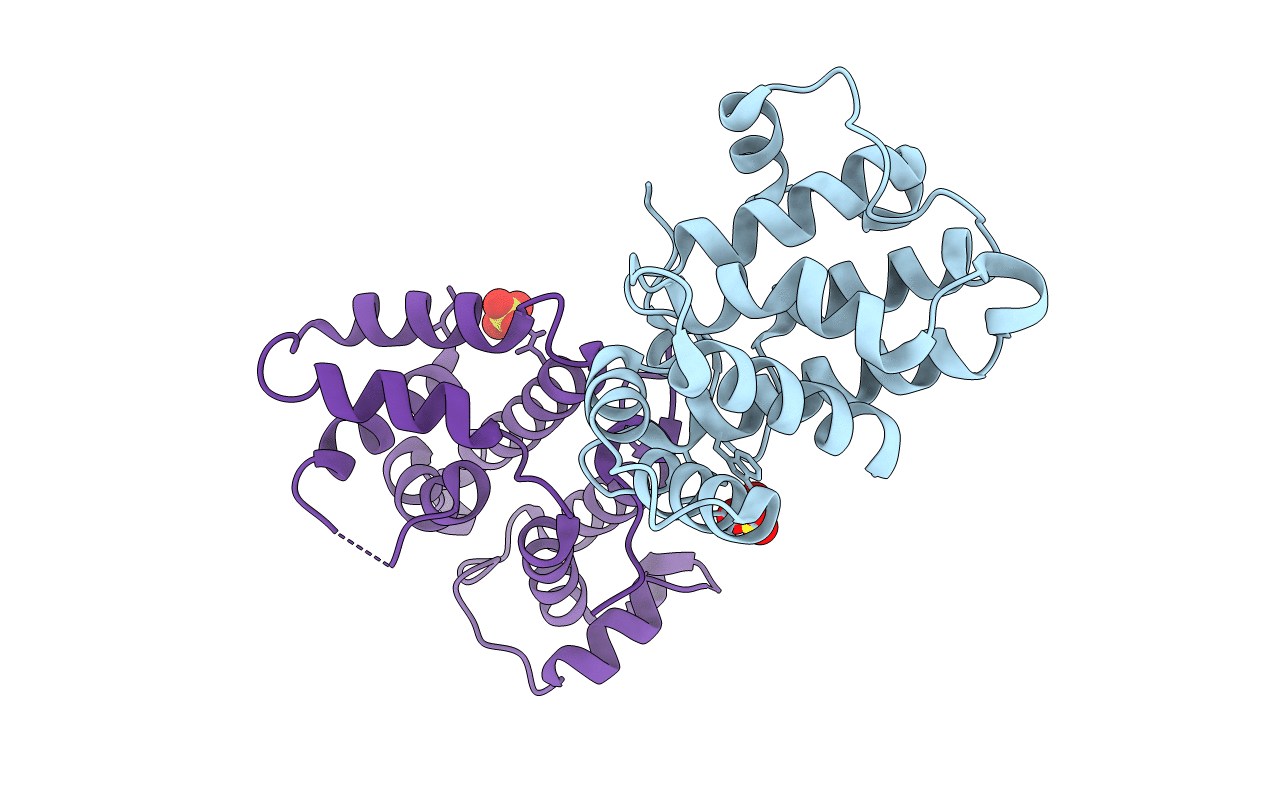 |
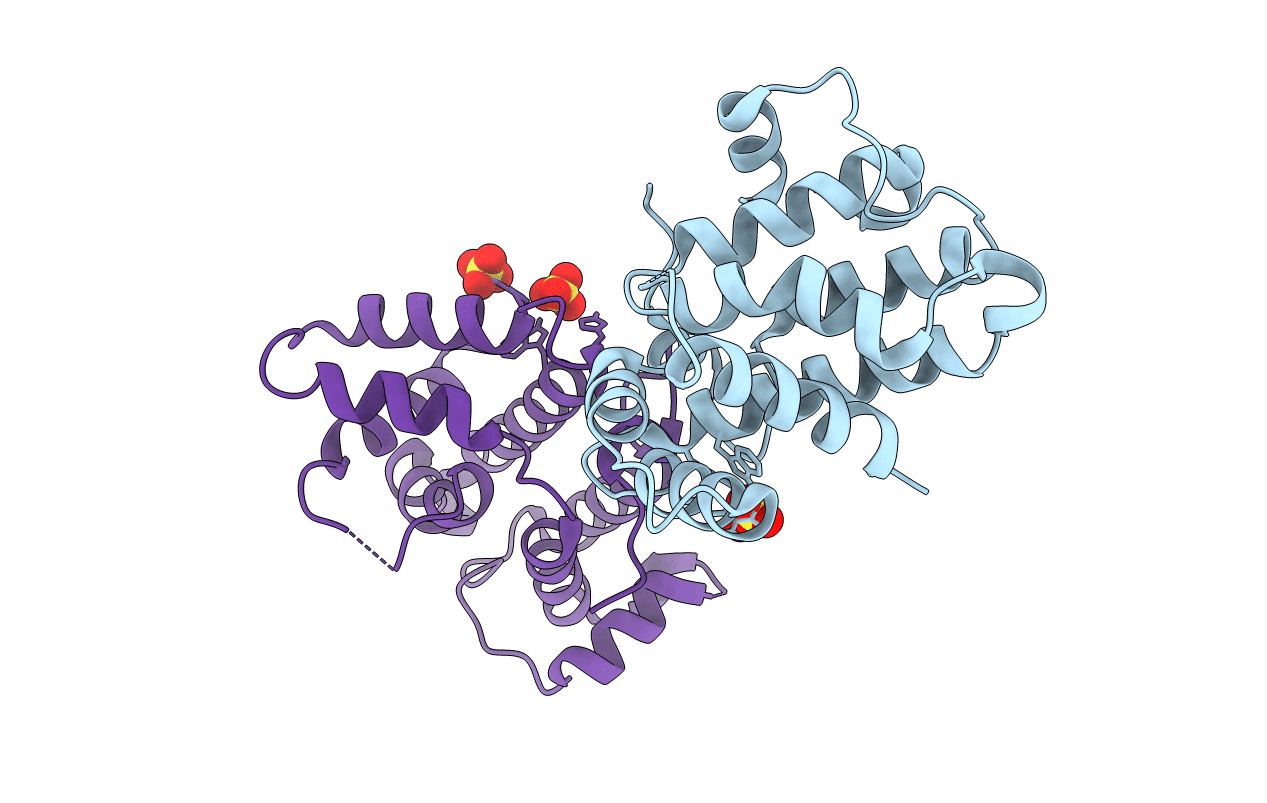
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-05-23 Classification: SIGNALING PROTEIN Ligands: SO4 |
 |
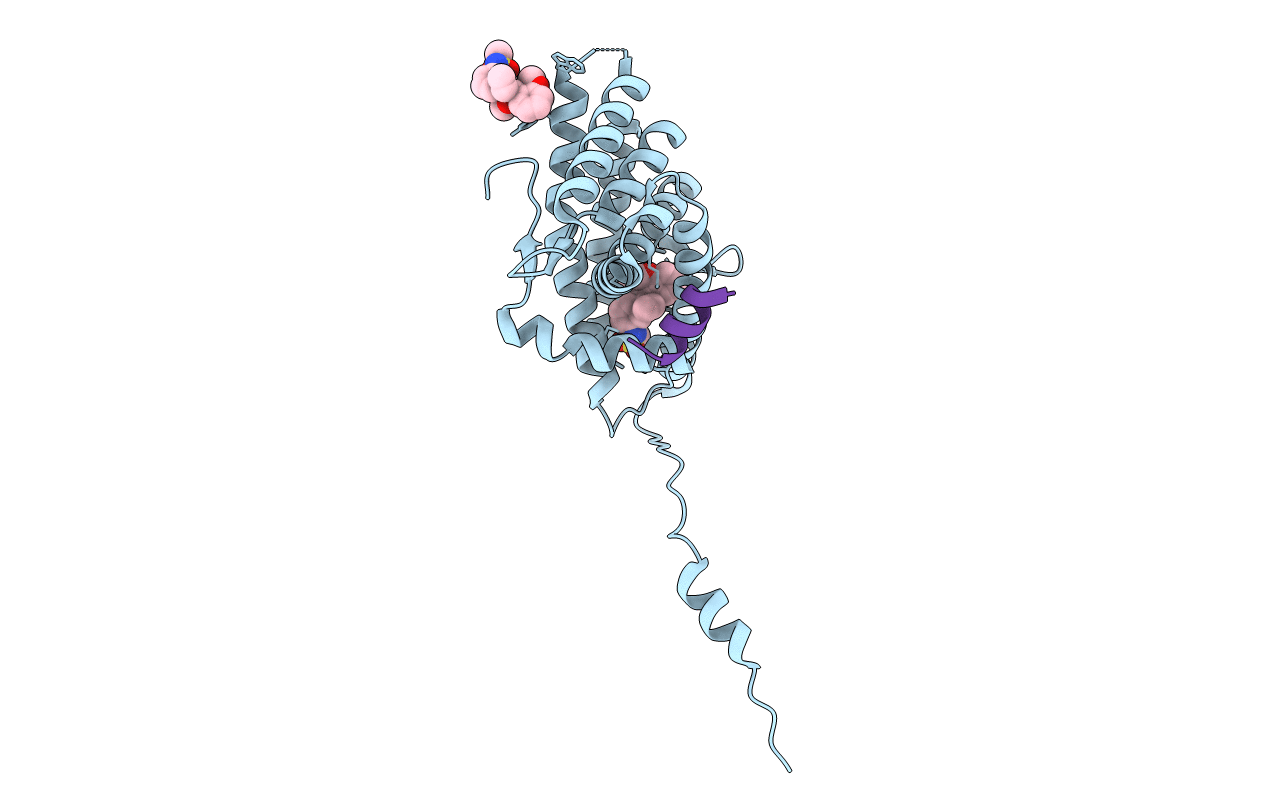
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-05-23 Classification: SIGNALING PROTEIN Ligands: SO4 |
 |
Crystal Structure Of The Glucocorticoid Receptor Ligand Binding Domain Bound To A Dibenzoxapine Sulfonamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2014-01-29 Classification: TRANSCRIPTION Ligands: LSJ |
 |
Solution Structure And Function Of A Conserved Protein Sp14.3 Encoded By An Essential Streptococcus Pneumoniae Gene
Organism: Streptococcus pneumoniae
Method: SOLUTION NMR Release Date: 2002-03-27 Classification: NUCLEIC ACID BINDING PROTEIN |

