Search Count: 160
 |
Organism: Penicillium steckii
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2025-03-05 Classification: OXIDOREDUCTASE Ligands: FMN, GOL, CL, CA |
 |
Crystal Structure Of Artificial Enzyme Lmrr_Paf Variant Rmh In Crystal Form 1
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2025-02-26 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of Artificial Enzyme Lmrr_Paf Variant Rmh In Crystal Form 2
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2025-02-26 Classification: DNA BINDING PROTEIN |
 |
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2025-02-26 Classification: DNA BINDING PROTEIN |
 |
Organism: Rhodococcus jostii rha1
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: FAD, B3P, MPD, MRD, CL |
 |
Crystal Structure Of Lmrr With V15 Replaced By Unnatural Amino Acid 4-Mercaptophenylalanine, Apo
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-12-25 Classification: METAL BINDING PROTEIN Ligands: HOH |
 |
Crystal Structure Of Lmrr With V15 Replaced By Unnatural Amino Acid 4-Mercaptophenylalanine, Au(I) Bound
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-12-25 Classification: METAL BINDING PROTEIN Ligands: AU |
 |
Organism: Rhodococcus jostii rha1
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2024-12-18 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Rhodococcus jostii rha1
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-12-18 Classification: OXIDOREDUCTASE Ligands: FAD, B3P, MPD, CL |
 |
Organism: Cyanobacterium tdx16
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: HEM, GOL, CA, MG |
 |
Organism: Oscillatoria princeps rmcb-10
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: FAD, CL |
 |
Crystal Structure Of T-Anethole Oxygenase From Stenotrophomonas Maltophilia
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2024-08-14 Classification: OXIDOREDUCTASE Ligands: HEM, GOL, CL |
 |
Crystal Structure Of Alcohol Dehydrogenase/Ketoreductase Variant From Thermus Thermophilus Apo Form
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-06-26 Classification: OXIDOREDUCTASE Ligands: SO4, GOL |
 |
Crystal Structure Of Alcohol Dehydrogenase/Ketoreductase Variant From Thermus Thermophilus Apo Form
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-06-26 Classification: OXIDOREDUCTASE Ligands: NAD, B3P, GOL |
 |
Organism: Bacillus tequilensis
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2024-05-08 Classification: OXIDOREDUCTASE Ligands: FMN, GOL, CL, MG |
 |
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-05-01 Classification: TRANSCRIPTION |
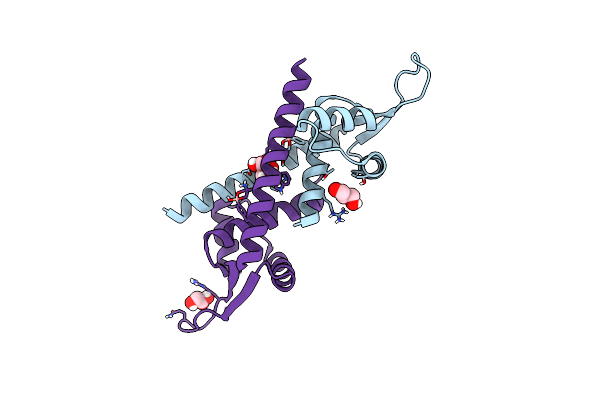 |
Engineered Lmrr Carrying A Cyclic Boronate Ester Formed Between Tris And P-Boronophenylalanine At Position 89
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2024-05-01 Classification: TRANSCRIPTION Ligands: GOL |
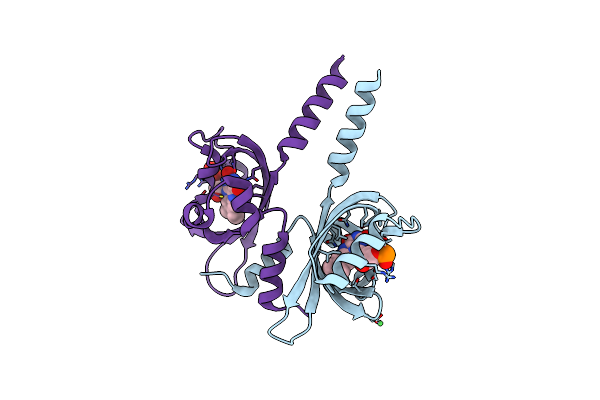 |
Crystal Structure Of Ppsb1-Lov Protein From Pseudomonas Putida With Covalent Fmn
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-11-29 Classification: SIGNALING PROTEIN Ligands: FMN, NI |
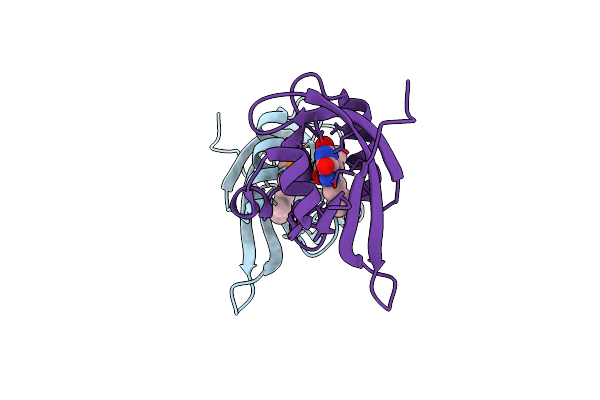 |
Crystal Structure Of Minisog Protein From Arabidopsis Thaliana With Covalent Fmn
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-11-29 Classification: SIGNALING PROTEIN Ligands: FMN |
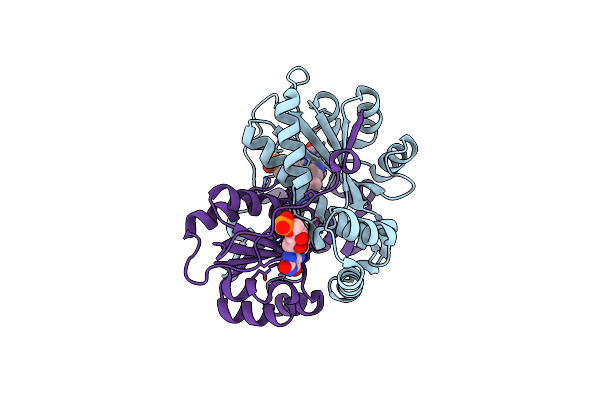 |
Crystal Structure Of Nitroreductase From Bacillus Tequilensis With Covalent Fmn
Organism: Bacillus tequilensis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-11-29 Classification: OXIDOREDUCTASE Ligands: FMN |

