Search Count: 60
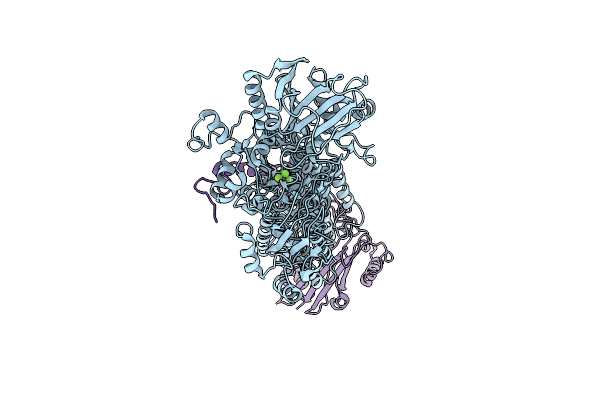 |
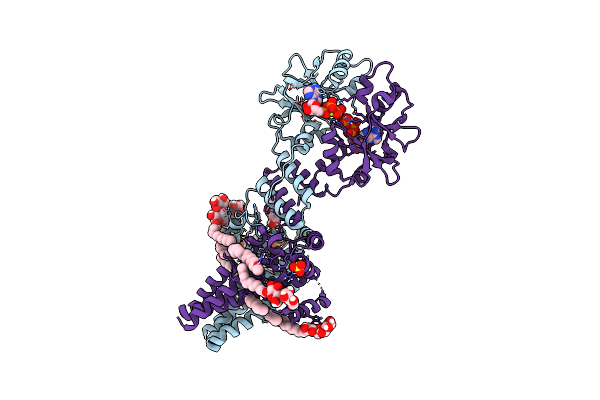
Cryo-Em Structure Of Na+,K+-Atpase Alpha2 From Artemia Salina In Cation-Free E2P Form
Organism: Artemia salina
Method: ELECTRON MICROSCOPY Release Date: 2023-11-29 Classification: TRANSPORT PROTEIN Ligands: ALF |
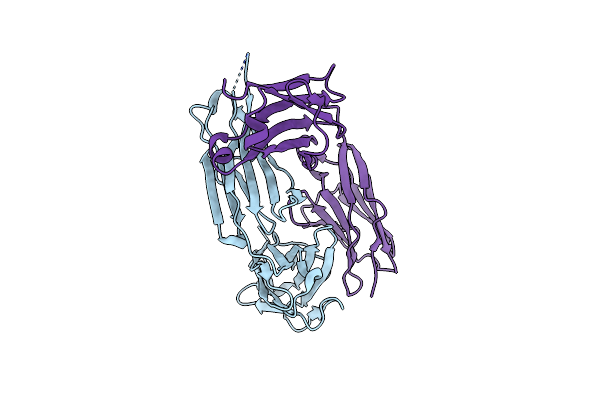 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2023-10-18 Classification: IMMUNE SYSTEM |
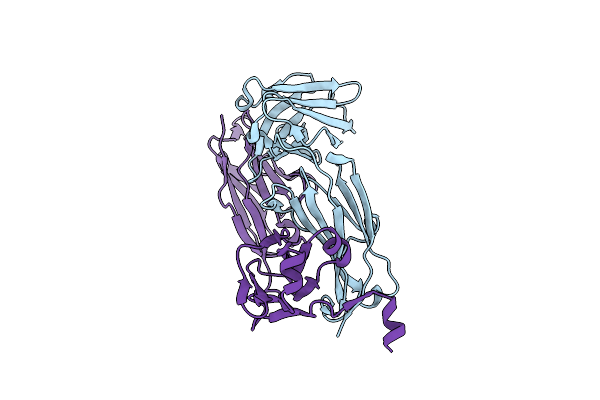 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2023-10-18 Classification: IMMUNE SYSTEM |
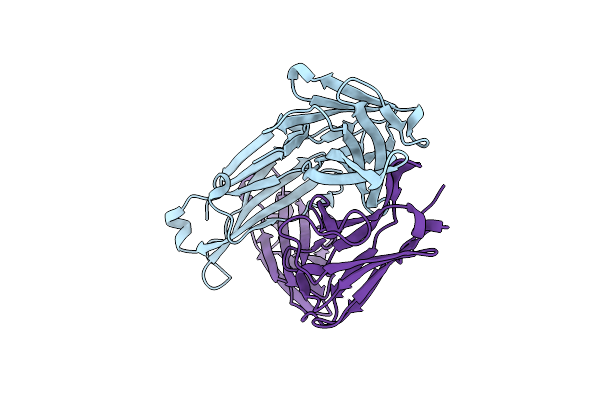 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2023-10-18 Classification: IMMUNE SYSTEM |
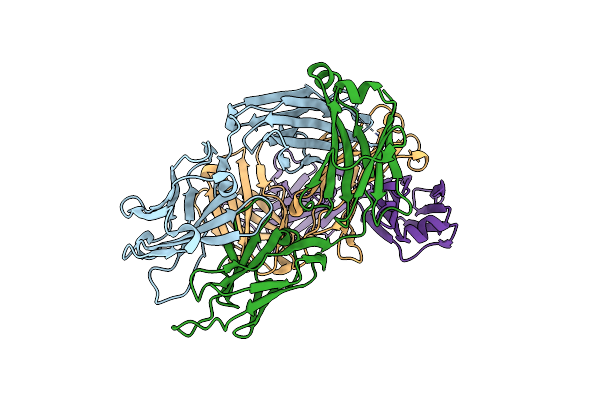 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2023-10-18 Classification: IMMUNE SYSTEM |
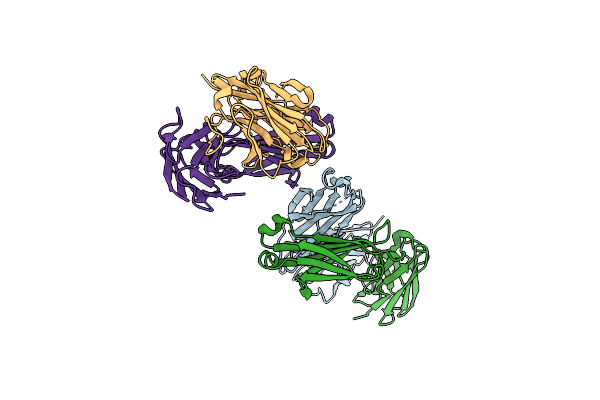 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2023-10-18 Classification: IMMUNE SYSTEM |
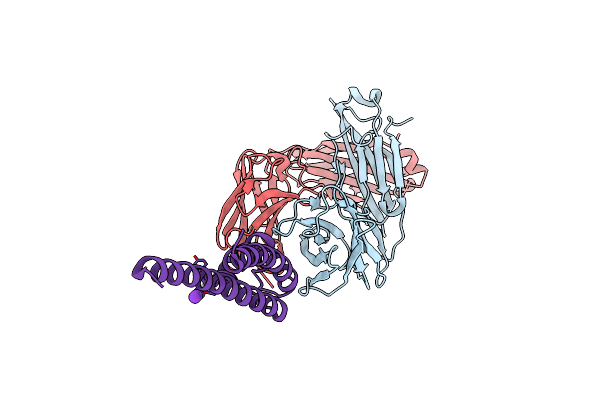 |
Organism: Mus musculus, Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2022-03-23 Classification: MEMBRANE PROTEIN Ligands: K |
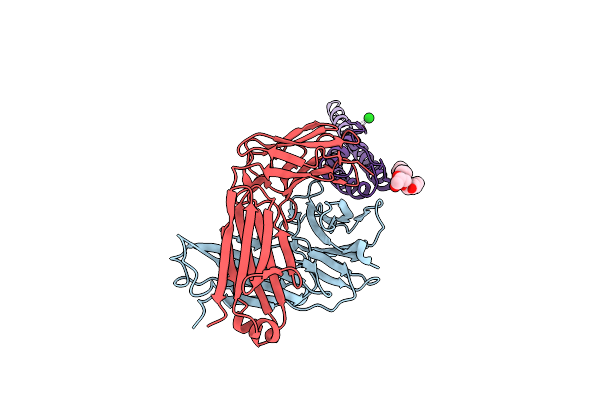 |
Organism: Mus musculus, Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2022-03-23 Classification: MEMBRANE PROTEIN Ligands: BA, DGA |
 |
Organism: Synthetic construct, Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2022-03-23 Classification: MEMBRANE PROTEIN Ligands: BA |
 |
Organism: Synthetic construct, Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2022-03-23 Classification: MEMBRANE PROTEIN |
 |
Organism: Synthetic construct, Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-03-23 Classification: MEMBRANE PROTEIN Ligands: K |
 |
Organism: Synthetic construct, Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2022-02-16 Classification: MEMBRANE PROTEIN Ligands: K |
 |
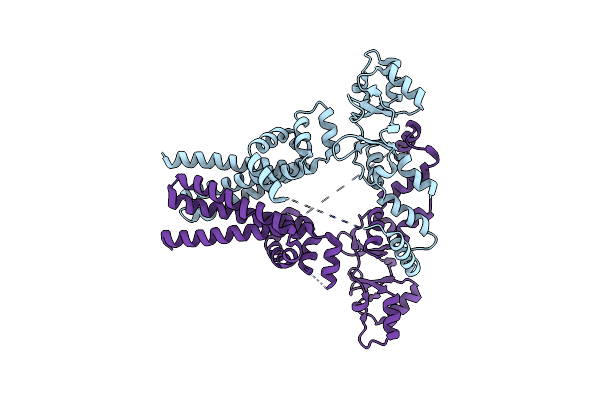
Crystal Structure Of An Archaeal Cnnm, Mtcorb, With C-Terminal Deletion In Complex With Mg2+-Atp
Organism: Methanoculleus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.26 Å Release Date: 2021-06-16 Classification: MEMBRANE PROTEIN Ligands: MG, SO4, UMQ, ATP, 1PE |
 |
Crystal Structure Of An Archaeal Cnnm, Mtcorb, R235L Mutant With C-Terminal Deletion
Organism: Methanoculleus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2021-06-16 Classification: MEMBRANE PROTEIN |
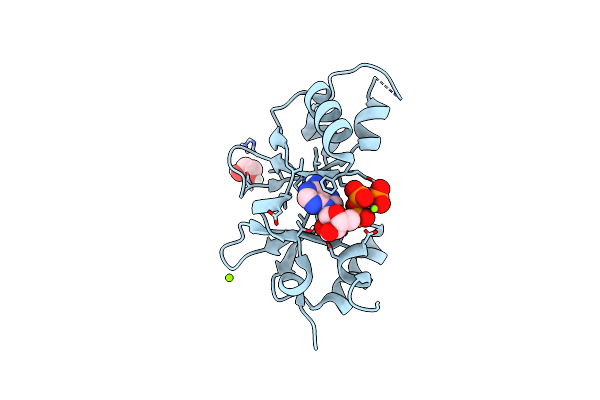 |
Crystal Structure Of An Archaeal Cnnm, Mtcorb, Cbs-Pair Domain In Complex With Mg2+-Atp
Organism: Methanoculleus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2021-06-16 Classification: MEMBRANE PROTEIN Ligands: EDO, MG, ATP |
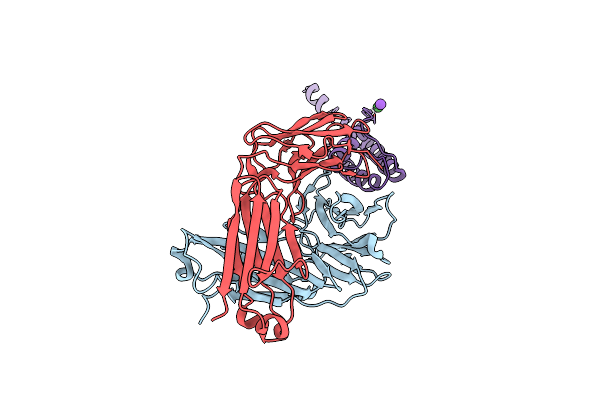 |
Organism: Rattus norvegicus, Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:3.24 Å Release Date: 2020-07-08 Classification: MEMBRANE PROTEIN Ligands: BA, K |
 |
Organism: Rattus norvegicus, Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2020-07-08 Classification: MEMBRANE PROTEIN Ligands: BA, K |
 |
Organism: Rattus norvegicus, Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:3.56 Å Release Date: 2020-07-08 Classification: MEMBRANE PROTEIN Ligands: BA, K |
 |
Organism: Rattus norvegicus, Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:3.64 Å Release Date: 2020-07-08 Classification: MEMBRANE PROTEIN Ligands: BA, K |
 |
Organism: Rattus norvegicus, Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:3.51 Å Release Date: 2020-07-08 Classification: MEMBRANE PROTEIN Ligands: BA, K |

