Search Count: 350
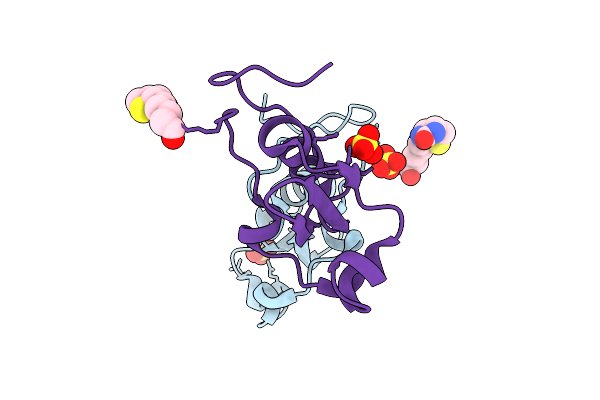 |
Structure Of Synthetic C-Terminal Biotinylated Schistosomin With The Point Mutation D88E From Biomphalaria Glabrata In P21 Space Group
Organism: Biomphalaria glabrata
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: TOXIN Ligands: BTN, SO4 |
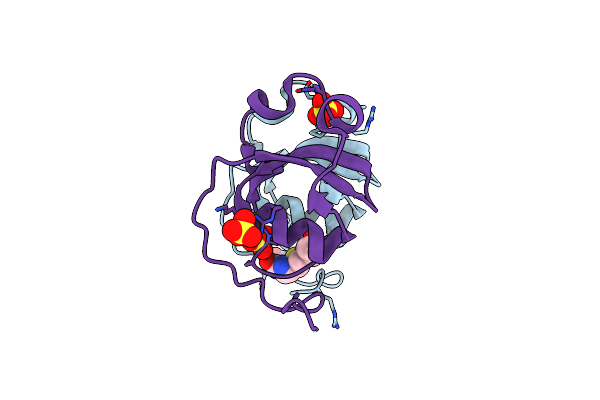 |
Structure Of Synthetic C-Terminal Biotinylated Schistosomin With The Point Mutation D88E From Biomphalaria Glabrata In C2 Space Group
Organism: Biomphalaria glabrata
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: TOXIN Ligands: BTN, SO4 |
 |
Organism: Candidatus heimdallarchaeota archaeon
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: MEMBRANE PROTEIN |
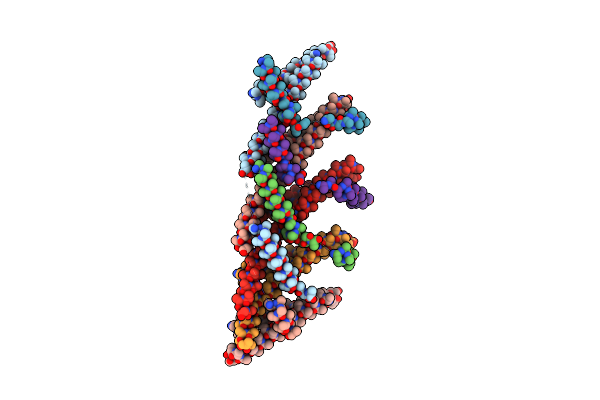 |
Organism: Candidatus heimdallarchaeota archaeon
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: MEMBRANE PROTEIN |
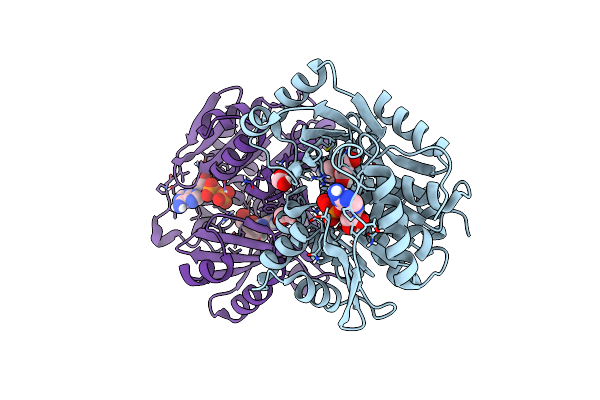 |
A Ternary Complex Of Plant Adenosine Kinase 1 From Moss Physcomitrella Patens (Ppadk1) With Adenosine And Adp
Organism: Physcomitrium patens
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2025-04-09 Classification: TRANSFERASE Ligands: ADP, ADN, NA, EDO |
 |
Crystal Structure Of Zea Mays Adenosine Kinase 3 (Zmadk3) In Complex With Ap5A
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2025-01-29 Classification: TRANSFERASE Ligands: AP5, EDO, PEG, CL |
 |
Organism: Dna molecule
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-01-22 Classification: DNA Ligands: MG, CL, HT1 |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2025-01-01 Classification: TRANSFERASE Ligands: SO4, ACT, GOL |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2025-01-01 Classification: TRANSFERASE Ligands: ACP, ACT, GOL |
 |
Organism: Nostoc punctiforme
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: LIPID BINDING PROTEIN |
 |
Cryo-Em Structure Of Vipp1-Deltah6_Aa1-219 Helical Filament With Lattice 2 (Vipp1-Deltah6_L2)
Organism: Nostoc punctiforme
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: LIPID BINDING PROTEIN |
 |
Cryo-Em Structure Of Vipp1-F197K/L200K Helical Filament With Lattice 1 (Vipp1-F197K/L200K_L1)
Organism: Nostoc punctiforme
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: LIPID BINDING PROTEIN |
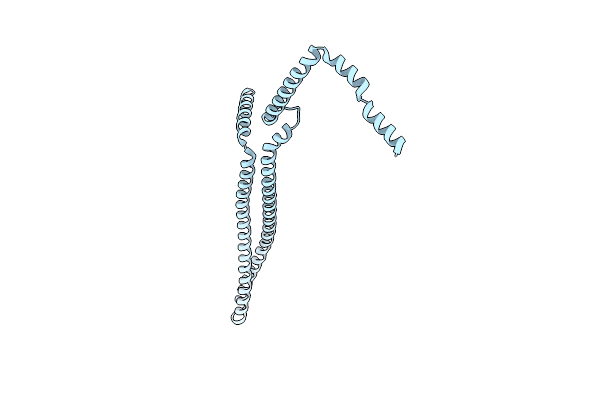 |
Cryo-Em Structure Of Vipp1-Deltah6_Aa1-219 Helical Filament With Lattice 3 (Vipp1-Deltah6_L3)
Organism: Nostoc punctiforme
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: LIPID BINDING PROTEIN |
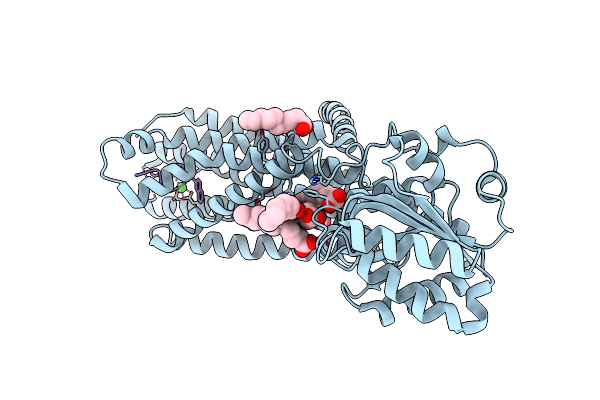 |
Crystal Structure Of The Melanocortin-4 Receptor (Mc4R) In Complex With S25
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-08-07 Classification: MEMBRANE PROTEIN Ligands: CA, OLA |
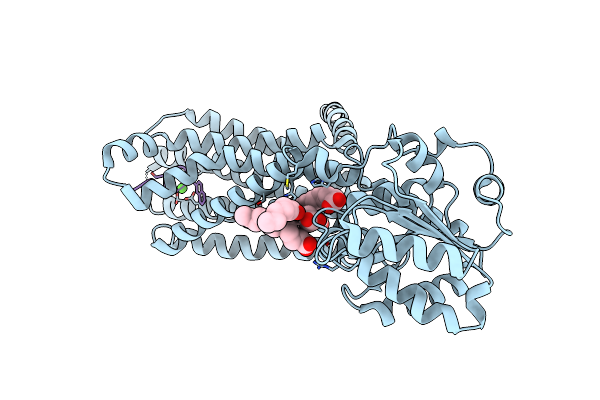 |
Crystal Structure Of The Melanocortin-4 Receptor (Mc4R) In Complex With S31
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2024-08-07 Classification: MEMBRANE PROTEIN Ligands: CA, OLA |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: LIPID BINDING PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: LIPID BINDING PROTEIN |
 |
Helical Reconstruction Of Yeast Eisosome Protein Pil1 Bound To Membrane Composed Of Lipid Mixture -Pip2/+Sterol (Dopc, Dope, Dops, Cholesterol 30:20:20:30)
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: LIPID BINDING PROTEIN |
 |
Helical Reconstruction Of Yeast Eisosome Protein Pil1 Bound To Membrane Composed Of Lipid Mixture +Pip2/-Sterol (Dopc, Dope, Dops, Pi(4,5)P2 50:20:20:10)
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: LIPID BINDING PROTEIN Ligands: I3P |
 |
Helical Reconstruction Of Yeast Eisosome Protein Pil1 Bound To Membrane Composed Of Lipid Mixture +Pip2/+Sterol (Dopc, Dope, Dops, Cholesterol, Pi(4,5)P2 35:20:20:15:10)
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: LIPID BINDING PROTEIN Ligands: I3P, P5S |

