Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Crystal Structure Of Periplasmic Domain Of Helicobacter Pylori Flil (Residues 81 To 183) (Crystal Form A)
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-03-23 Classification: MOTOR PROTEIN |
 |
Crystal Structure Of Periplasmic Domain Of Helicobacter Pylori Flil (Residues 81 To 183) (Crystal Form B)
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-03-23 Classification: MOTOR PROTEIN |
 |
Crystal Structure Of Periplasmic Domain Of Helicobacter Pylori Flil (Residues 81 To 183) (Crystal Form C)
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-03-23 Classification: MOTOR PROTEIN |
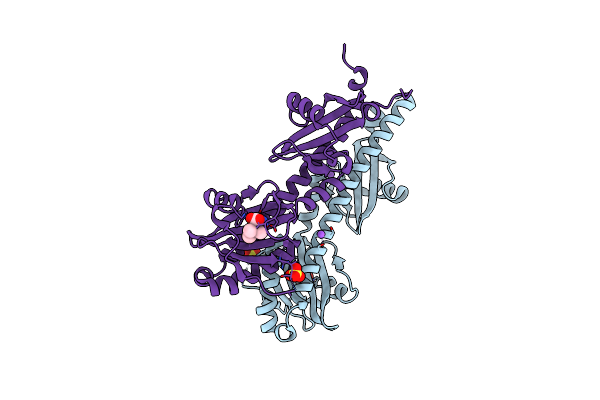 |
Crystal Structure Of Ligand-Binding Domain Of Campylobacter Jejuni Chemoreceptor Tlp3 In Complex With 4-Methylisoleucine
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2020-05-20 Classification: SIGNALING PROTEIN Ligands: SKG, SO4, NA, CL |
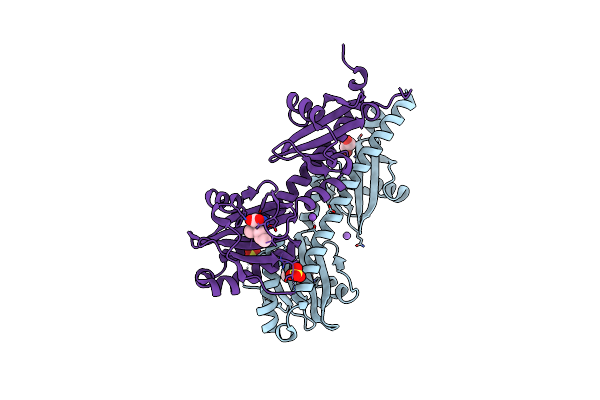 |
Crystal Structure Of Ligand-Binding Domain Of Campylobacter Jejuni Chemoreceptor Tlp3 In Complex With Beta-Methylnorleucine
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2020-05-20 Classification: SIGNALING PROTEIN Ligands: SKJ, SO4, CL, GOL, NA |
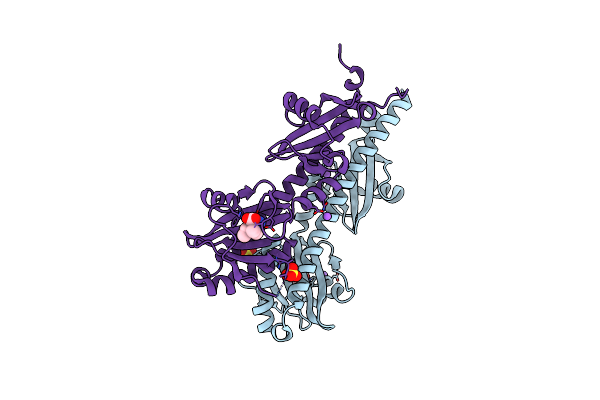 |
Crystal Structure Of Ligand-Binding Domain Of Campylobacter Jejuni Chemoreceptor Tlp3 In Complex With 3-Methylisoleucine
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2020-05-20 Classification: SIGNALING PROTEIN Ligands: I2M, SO4, CL, NA |
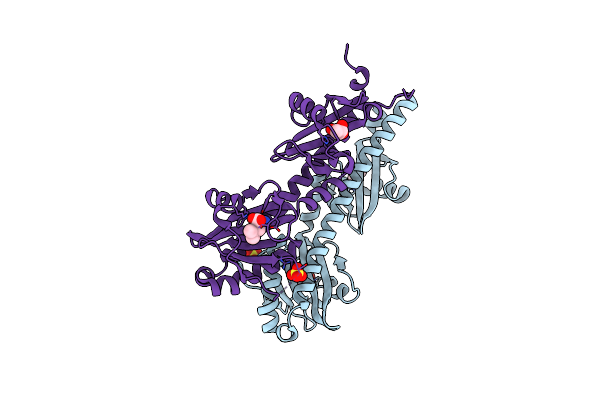 |
Crystal Structure Of Ligand-Binding Domain Of Campylobacter Jejuni Chemoreceptor Tlp3 In Complex With L-Leucine
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-05-20 Classification: SIGNALING PROTEIN Ligands: LEU, GOL, SO4 |
 |
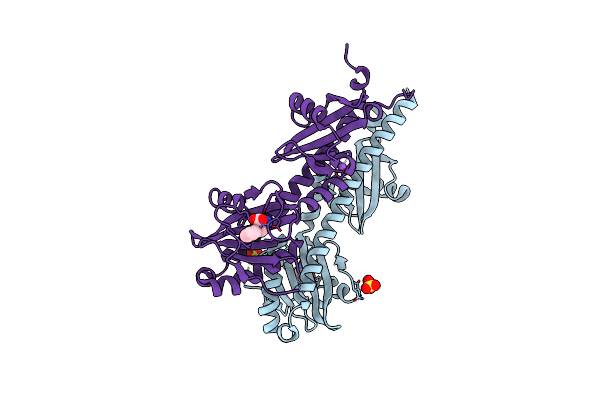
Crystal Structure Of Ligand-Binding Domain Of Campylobacter Jejuni Chemoreceptor Tlp3 In Complex With L-Norvaline
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-05-20 Classification: SIGNALING PROTEIN Ligands: NVA, SO4, GOL |
 |
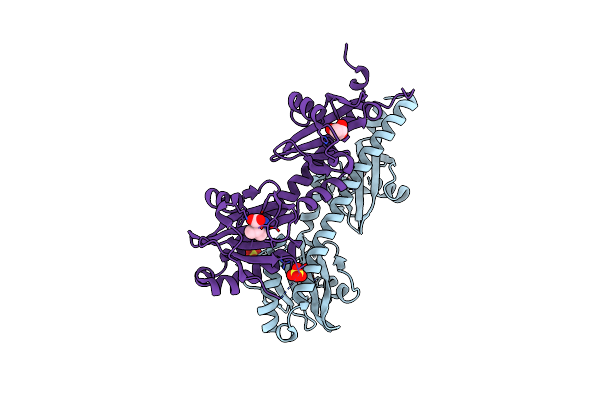
Crystal Structure Of Ligand-Binding Domain Of Campylobacter Jejuni Chemoreceptor Tlp3 In Complex With L-Phenylalanine
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2020-05-20 Classification: SIGNALING PROTEIN Ligands: PHE, SO4, CL, NA |
 |
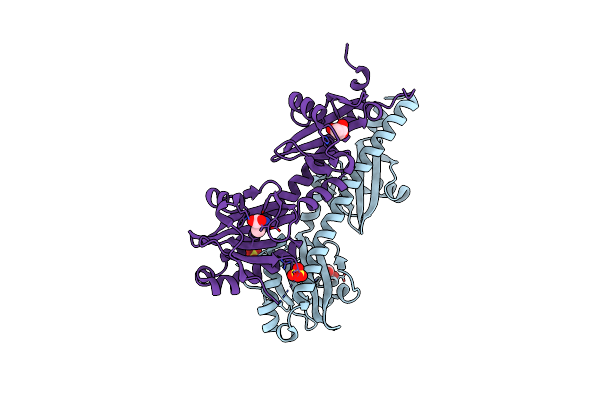
Crystal Structure Of Ligand-Binding Domain Of Campylobacter Jejuni Chemoreceptor Tlp3 In Complex With L-Valine
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-05-20 Classification: SIGNALING PROTEIN Ligands: VAL, GOL, SO4 |
 |
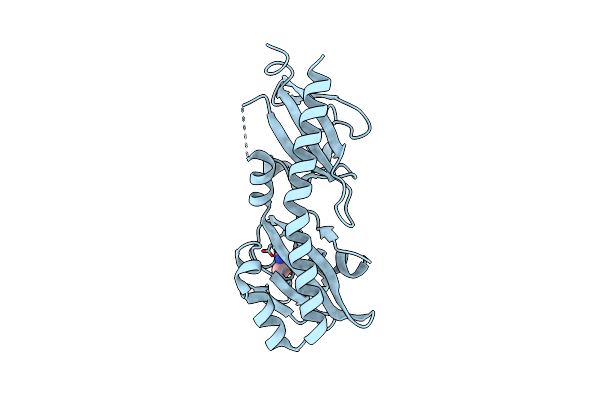
Crystal Structure Of Ligand-Binding Domain Of Campylobacter Jejuni Chemoreceptor Tlp3 In Complex With L-Alanine
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2020-05-20 Classification: SIGNALING PROTEIN Ligands: ALA, GOL, SO4, CL |
 |
Crystal Structure Of Ligand-Binding Domain Of Pseudomonas Fluorescens Chemoreceptor Ctaa In Complex With L-Alanine
Organism: Pseudomonas fluorescens (strain pf0-1)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-03-18 Classification: SIGNALING PROTEIN Ligands: ALA |
 |
Crystal Structure Of Ligand-Binding Domain Of Pseudomonas Fluorescens Chemoreceptor Ctaa In Complex With L-Isoleucine
Organism: Pseudomonas fluorescens (strain pf0-1)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-03-18 Classification: SIGNALING PROTEIN Ligands: ILE |
 |
Crystal Structure Of Ligand-Binding Domain Of Pseudomonas Fluorescens Chemoreceptor Ctaa In Complex With L-Leucine
Organism: Pseudomonas fluorescens (strain pf0-1)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-03-18 Classification: SIGNALING PROTEIN Ligands: LEU |
 |
Crystal Structure Of Ligand-Binding Domain Of Pseudomonas Fluorescens Chemoreceptor Ctaa In Complex With L-Serine
Organism: Pseudomonas fluorescens (strain pf0-1)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-03-18 Classification: SIGNALING PROTEIN Ligands: SER |
 |
Crystal Structure Of Ligand-Binding Domain Of Pseudomonas Fluorescens Chemoreceptor Ctaa
Organism: Pseudomonas fluorescens (strain pf0-1)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-03-18 Classification: SIGNALING PROTEIN Ligands: ALA |
 |
Crystal Structure Of Ligand-Binding Domain Of Pseudomonas Fluorescens Chemoreceptor Ctaa In Complex With L-Valine
Organism: Pseudomonas fluorescens (strain pf0-1)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-03-18 Classification: SIGNALING PROTEIN Ligands: VAL, NA, CL |
 |
Crystal Structure Of Ligand-Binding Domain Of Pseudomonas Fluorescens Chemoreceptor Ctaa In Complex With L-Proline
Organism: Pseudomonas fluorescens (strain pf0-1)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-03-18 Classification: SIGNALING PROTEIN Ligands: PRO, CL |
 |
Abc Transporter-Associated Periplasmic Binding Protein Dppa From Helicobacter Pylori In Complex With Peptide Stsa
Organism: Helicobacter pylori ss1, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2019-08-21 Classification: Periplasmic peptide binding protein |
 |
Abc Transporter-Associated Periplasmic Binding Protein Dppa From Helicobacter Pylori
Organism: Helicobacter pylori, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-08-21 Classification: TRANSPORT PROTEIN |