Search Count: 13
 |
Organism: Streptococcus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-12-12 Classification: METAL BINDING PROTEIN Ligands: NI |
 |
Organism: Streptococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-04-04 Classification: METAL BINDING PROTEIN Ligands: ZN, MPD, NA, SO4, EDO, GOL |
 |
Organism: Streptococcus sp. group g
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2018-04-04 Classification: METAL BINDING PROTEIN Ligands: ZN, ACT, EDO, MPD, MRD, CL, SO4 |
 |
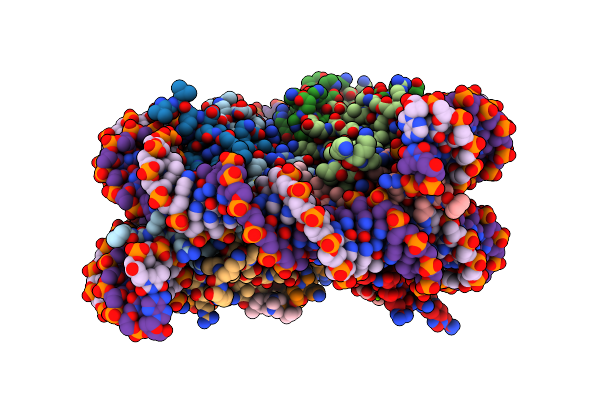
Nucleosome Core Particle Containing Adducts Of Ruthenium(Ii)-Toluene Pta Complex
Organism: Xenopus laevis, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2016-09-14 Classification: STRUCTURAL PROTEIN/DNA Ligands: SO4, MG, RAX |
 |
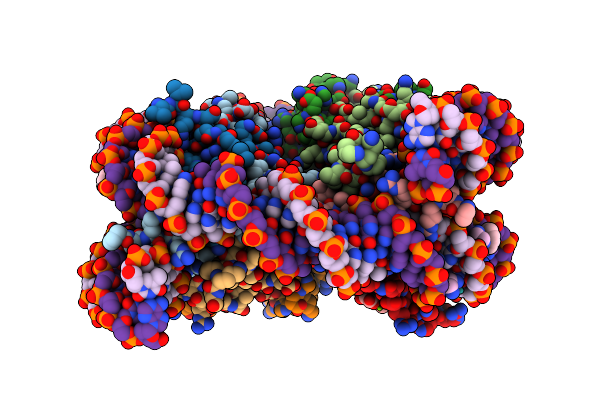
Nucleosome Core Particle Containing Adducts Of Gold(I)-Triethylphosphane And Ruthenium(Ii)-Toluene Pta Complexes
Organism: Xenopus laevis, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-09-14 Classification: STRUCTURAL PROTEIN/DNA Ligands: AUF, SO4, MG, RAX |
 |
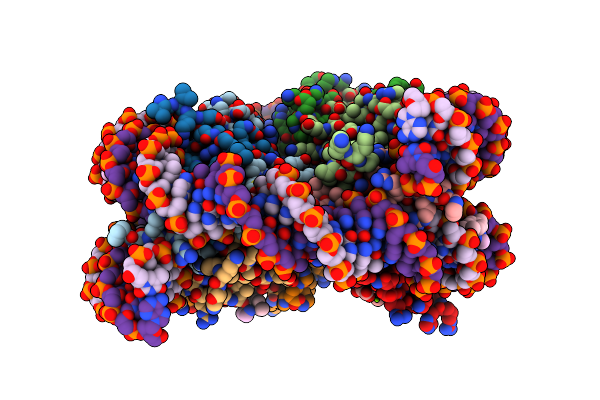
Nucleosome Core Particle With Adducts From The Anticancer Compound, [(Eta6-5,8,9,10-Tetrahydroanthracene)Ru(Ethylenediamine)Cl][Pf6]
Organism: Xenopus laevis, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-06-01 Classification: STRUCTURAL PROTEIN/DNA Ligands: RUH, SO4, MG |
 |
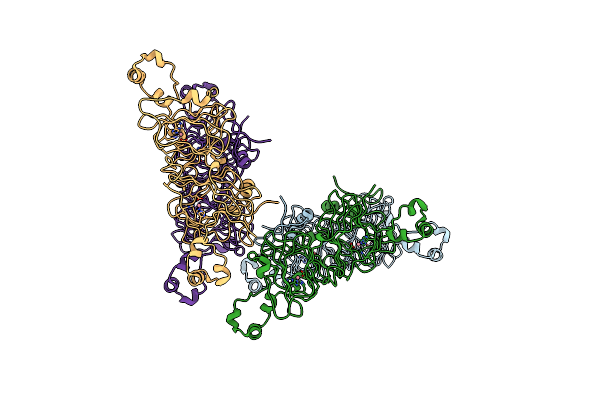
Nucleosome Core Particle Containing (Eta6-P-Cymene)-(1, 2-Ethylenediamine)-Ruthenium
Organism: Xenopus laevis, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2014-03-26 Classification: STRUCTURAL PROTEIN/DNA Ligands: HRU, SO4, MG |
 |
Assigning The Epr Fine Structure Parameters Of The Mn(Ii) Centers In Bacillus Subtilis Oxalate Decarboxylase By Site-Directed Mutagenesis And Dft/Mm Calculations
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-03-05 Classification: LYASE Ligands: CO |
 |
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-10-09 Classification: UNKNOWN FUNCTION Ligands: 1PG, 6NA, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2012-10-03 Classification: ISOMERASE Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-10-03 Classification: ISOMERASE Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-10-03 Classification: ISOMERASE Ligands: SO4 |
 |
Structure Of Human Glutathione Transferase Pi Class In Complex With Ethacraplatin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2011-05-11 Classification: TRANSFERASE Ligands: PT, MES, EAA, CA, CL |

