Search Count: 8
 |
Structure Of Sasg (Type Ii) (Residues 165-421) From Staphylococcus Aureus Mw2
Organism: Staphylococcus aureus subsp. aureus mw2
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2024-05-08 Classification: CELL ADHESION Ligands: SO4, CA, NA, ZN |
 |
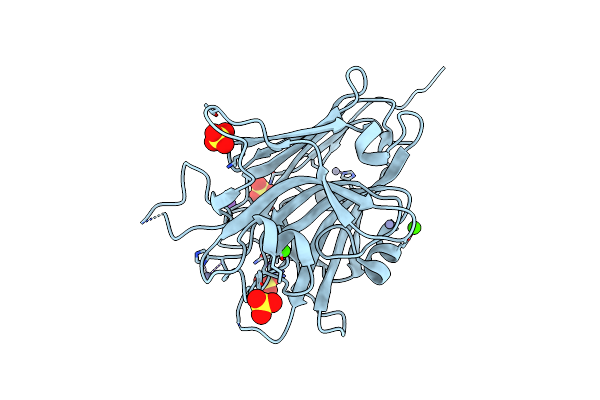
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2022-06-01 Classification: HORMONE |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-10-21 Classification: VIRAL PROTEIN |
 |
X-Ray Structure Of The Dimeric Cytochrome Bc1 Complex From The Soil Bacterium Paracoccus Denitrificans At 2.7 Angstrom Resolution
Organism: Paracoccus denitrificans
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2011-10-26 Classification: OXIDOREDUCTASE Ligands: HEM, SMA, HEC, FES |
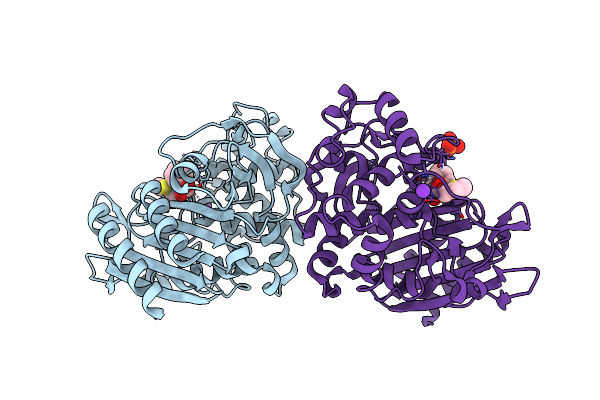 |
Ampc Beta-Lactamase N289A Mutant In Complex With A Boronic Acid Deacylation Transition State Analog Compound Sm3
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.07 Å Release Date: 2006-03-28 Classification: HYDROLASE Ligands: K, SM3, PO4 |
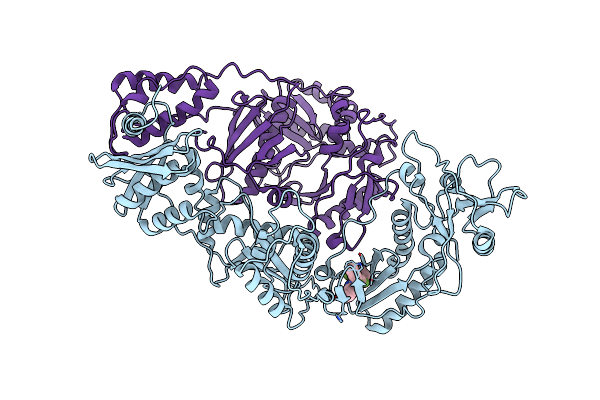 |
Crystal Structure Of Hiv-1 Reverse Transcriptase (Rt) In Complex With Thr-50
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2005-11-01 Classification: TRANSFERASE Ligands: T50 |
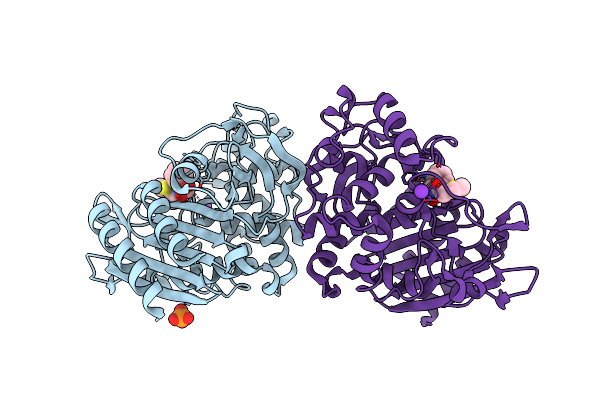 |
Structure Of N289A Mutant Of Ampc In Complex With Sm3, A Phenylglyclboronic Acid Bearing The Cephalothin R1 Side Chain
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2004-02-24 Classification: HYDROLASE Ligands: PO4, K, SM3 |
 |
Structure Of N289A Mutant Of Ampc In Complex With Sm2, Carboxyphenylglycylboronic Acid Bearing The Cephalothin R1 Side Chain
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2004-02-24 Classification: HYDROLASE Ligands: PO4, K, SM2 |

