Search Count: 21
 |
Cryo-Em Structure Of The Glucose-Specific Pts Transporter Iicb From E. Coli In An Intermediate State
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-04-02 Classification: TRANSPORT PROTEIN Ligands: LMT |
 |
Cryo-Em Structure Of The Glucose-Specific Pts Transporter Iicb From E. Coli In The Inward-Facing Conformation
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-09-25 Classification: TRANSPORT PROTEIN Ligands: BGC |
 |
Cryo-Em Structure Of The Glucose-Specific Pts Transporter Iicb From E. Coli In The Inward- And Outward-Facing Conformation
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-09-25 Classification: TRANSPORT PROTEIN Ligands: BGC |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: MEMBRANE PROTEIN Ligands: FES, HEM, U10, I7Y, HEC, CU, CA |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: MEMBRANE PROTEIN Ligands: CU, CA, HEM, FES, I7Y, U10, HEC |
 |
Organism: Schizosaccharomyces pombe
Method: ELECTRON MICROSCOPY Release Date: 2023-03-01 Classification: MEMBRANE PROTEIN Ligands: HEA, CU, MG, CA, PEF, CUA, ZN |
 |
Nmr Structural Analysis Of Yeast Cox13 Reveals Its C-Terminus In Interaction With Atp
Organism: Saccharomyces cerevisiae s288c
Method: SOLUTION NMR Release Date: 2021-05-19 Classification: STRUCTURAL PROTEIN |
 |
Crystal Structure Of I107E Cub Myoglobin (I107E L29H F43H Sperm Whale Myoglobin)
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2021-02-03 Classification: OXYGEN TRANSPORT Ligands: HEM, PG4, EDO, SO4, NA |
 |
Crystal Structure Of I107E F33Y Cub Myoglobin (I107E F33Y L29H F43H Sperm Whale Myoglobin)
Organism: Physeter macrocephalus
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2021-02-03 Classification: OXYGEN TRANSPORT Ligands: HEM |
 |
Crystal Structure Of Oxy-I107E Cub Myoglobin (I107E L29H F43H Sperm Whale Myoglobin; Partial Occupancy)
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2021-02-03 Classification: OXYGEN TRANSPORT Ligands: HEM, PER, CL |
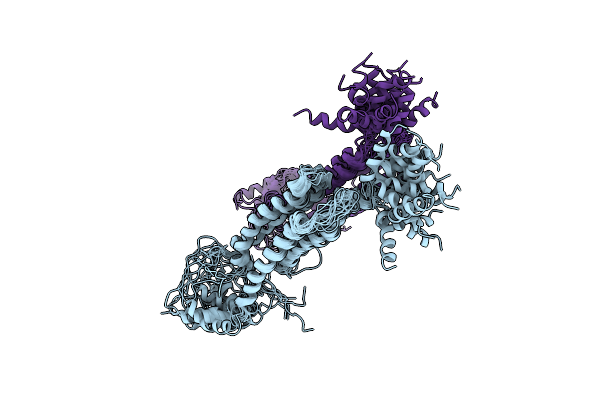 |
Nmr Structure And Dynamics Studies Of Yeast Respiratory Super-Complex Factor 2 In Micelles
Organism: Saccharomyces cerevisiae s288c
Method: SOLUTION NMR Release Date: 2020-10-07 Classification: STRUCTURAL PROTEIN |
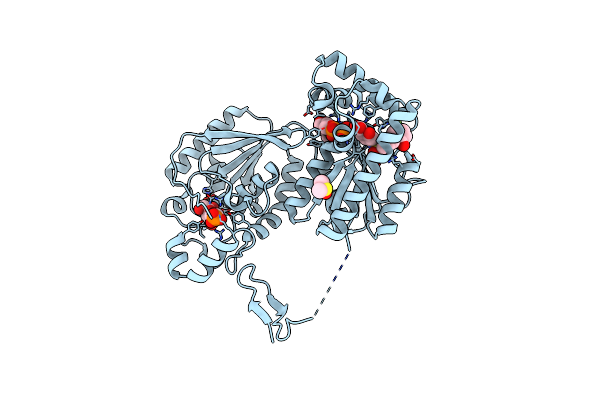 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2018-11-07 Classification: ANTITUMOR PROTEIN Ligands: DMS, F6P, BWW, ADP |
 |
Structure Of A Functional Obligate Respiratory Supercomplex From Mycobacterium Smegmatis
Organism: Mycobacterium smegmatis mc2 155, Mycobacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: ELECTRON MICROSCOPY Release Date: 2018-11-07 Classification: ELECTRON TRANSPORT Ligands: FES, CDL, MQ9, CU, HAS, HEC, HEM |
 |
Organism: Neisseria meningitidis (strain alpha14)
Method: X-RAY DIFFRACTION Resolution:4.20 Å Release Date: 2018-03-14 Classification: OXIDOREDUCTASE Ligands: HEM, CA, FE |
 |
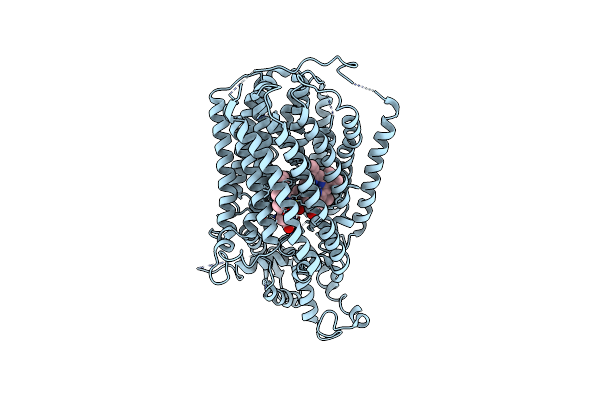
Solution Structure Of Detergent-Solubilized Rcf1, A Yeast Mitochondrial Inner Membrane Protein Involved In Respiratory Complex Iii/Iv Supercomplex Formation
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2018-02-28 Classification: MEMBRANE PROTEIN |
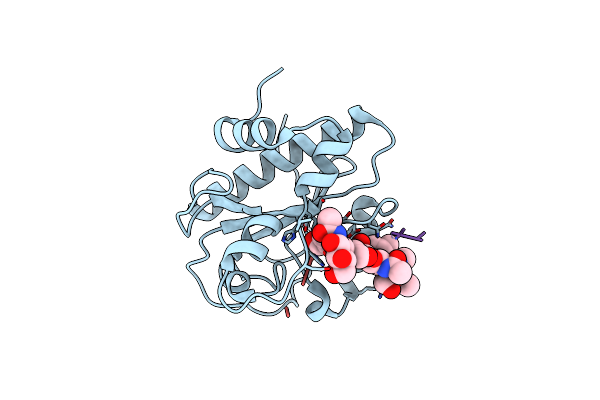 |
Catalytic Domain Of Lyta, The Major Autolysin Of Streptococcus Pneumoniae, (C60A, H133A, C136A Mutant) Complexed With Peptidoglycan Fragment
Organism: Streptococcus pneumoniae serotype 4, Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2016-06-15 Classification: HYDROLASE |
 |
Catalytic Amidase Domain Of The Major Autolysin Lyta From Streptococcus Pneumaniae
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2014-01-29 Classification: HYDROLASE Ligands: ZN, EDO |
 |
Crystal Structure Of The C-Teminal Choline-Binding Domain Of The Streptococcus Pneumoniae Prophage Lyta
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-01-29 Classification: HYDROLASE Ligands: CHT |
 |
Crystal Structure Of The F1-Atpase From The Thermoalkaliphilic Bacterium Bacillus Sp. Ta2.A1
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2007-08-21 Classification: HYDROLASE |
 |
Crystal Structure Of The Carboxyltransferase Domain Of The Oxaloacetate Decarboxylase Na+ Pump From Vibrio Cholerae
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2006-12-26 Classification: LYASE Ligands: ZN |

