Search Count: 402
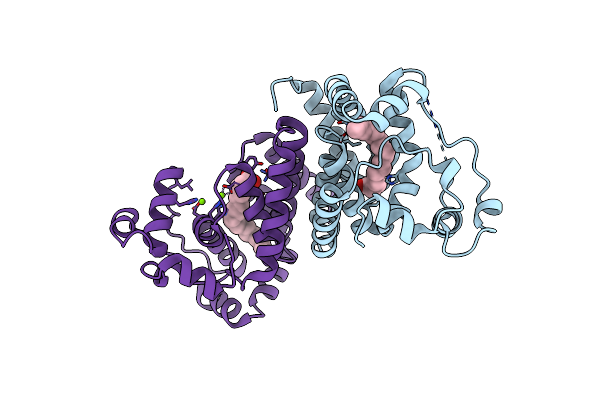 |
Crystal Structure Of The Alr8543 Protein In Complex With Oleic Acid And Magnesium Ion From Nostoc Sp. Pcc 7120, Northeast Structural Genomics Consortium Target Nsr141
Organism: Nostoc sp. (strain pcc 7120 / sag 25.82 / utex 2576)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-07-25 Classification: LIPID BINDING PROTEIN Ligands: OLA, MG, CL |
 |
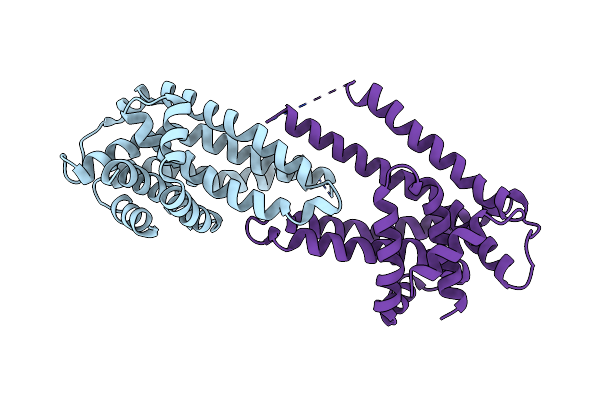
Organism: Pseudomonas denitrificans (nomen rejiciendum)
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2018-05-23 Classification: TRANSPORT PROTEIN Ligands: BOG |
 |
Organism: Pseudomonas fragi a22
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2018-05-16 Classification: TRANSPORT PROTEIN |
 |
Organism: Cupriavidus metallidurans (strain atcc 43123 / dsm 2839 / nbrc 102507 / ch34)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2016-02-17 Classification: TRANSFERASE Ligands: ZN, DSL, MPG, CL, PO4, PC, EPE |
 |
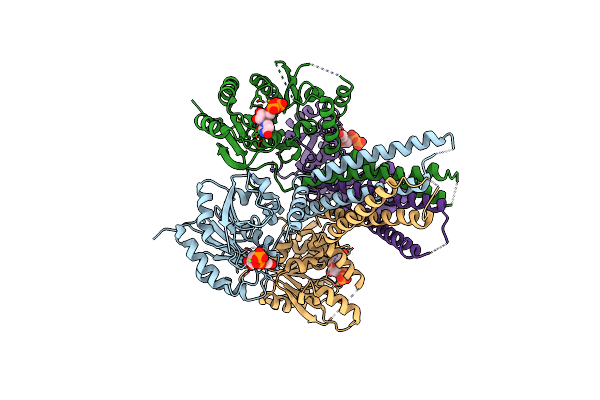
Crystal Structure Of Arnt From Cupriavidus Metallidurans Bound To Undecaprenyl Phosphate
Organism: Cupriavidus metallidurans (strain atcc 43123 / dsm 2839 / nbrc 102507 / ch34)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-02-17 Classification: TRANSFERASE Ligands: MPG, EPE, PO4, CL, 5TR |
 |
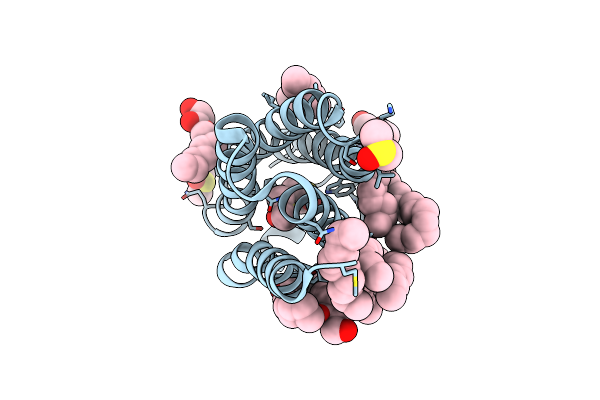
Structure Of The Polyisoprenyl-Phosphate Glycosyltransferase Gtrb (F215A Mutant)
Organism: Synechocystis sp. (strain pcc 6803 / kazusa)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-01-06 Classification: TRANSFERASE Ligands: UDP, MG |
 |
Organism: Synechocystis sp. (strain pcc 6803 / kazusa)
Method: X-RAY DIFFRACTION Resolution:3.19 Å Release Date: 2016-01-06 Classification: TRANSFERASE Ligands: UDP, MG |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-04-22 Classification: MEMBRANE PROTEIN Ligands: MPG, DMS |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:4.10 Å Release Date: 2015-02-11 Classification: MEMBRANE PROTEIN |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2015-02-11 Classification: MEMBRANE PROTEIN Ligands: MPG, LMU, CAC, PGE |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:3.49 Å Release Date: 2015-01-28 Classification: MEMBRANE PROTEIN Ligands: PKA |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-01-28 Classification: MEMBRANE PROTEIN Ligands: MPG |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-01-28 Classification: MEMBRANE PROTEIN Ligands: MPG, DMS |
 |
Crystal Structure Of A Bacterial Bestrophin Homolog From Klebsiella Pneumoniae By Zn-Sad Phasing
Organism: Klebsiella pneumoniae uhkpc96
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2014-10-01 Classification: MEMBRANE PROTEIN Ligands: ZN |
 |
Crystal Structure Of A Bacterial Bestrophin Homolog From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae uhkpc96
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-10-01 Classification: MEMBRANE PROTEIN Ligands: ZN |
 |
Crystal Structure Of An Exopolyphosphatase-Related Protein From Bacteroides Fragilis. Northeast Structural Genomics Target Bfr192
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2014-06-25 Classification: UNKNOWN FUNCTION Ligands: PO4, NA |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2014-06-04 Classification: Membrance Protein |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-06-04 Classification: Membrance Protein |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2014-06-04 Classification: Membrance Protein |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2014-06-04 Classification: Membrance Protein |

