Search Count: 34
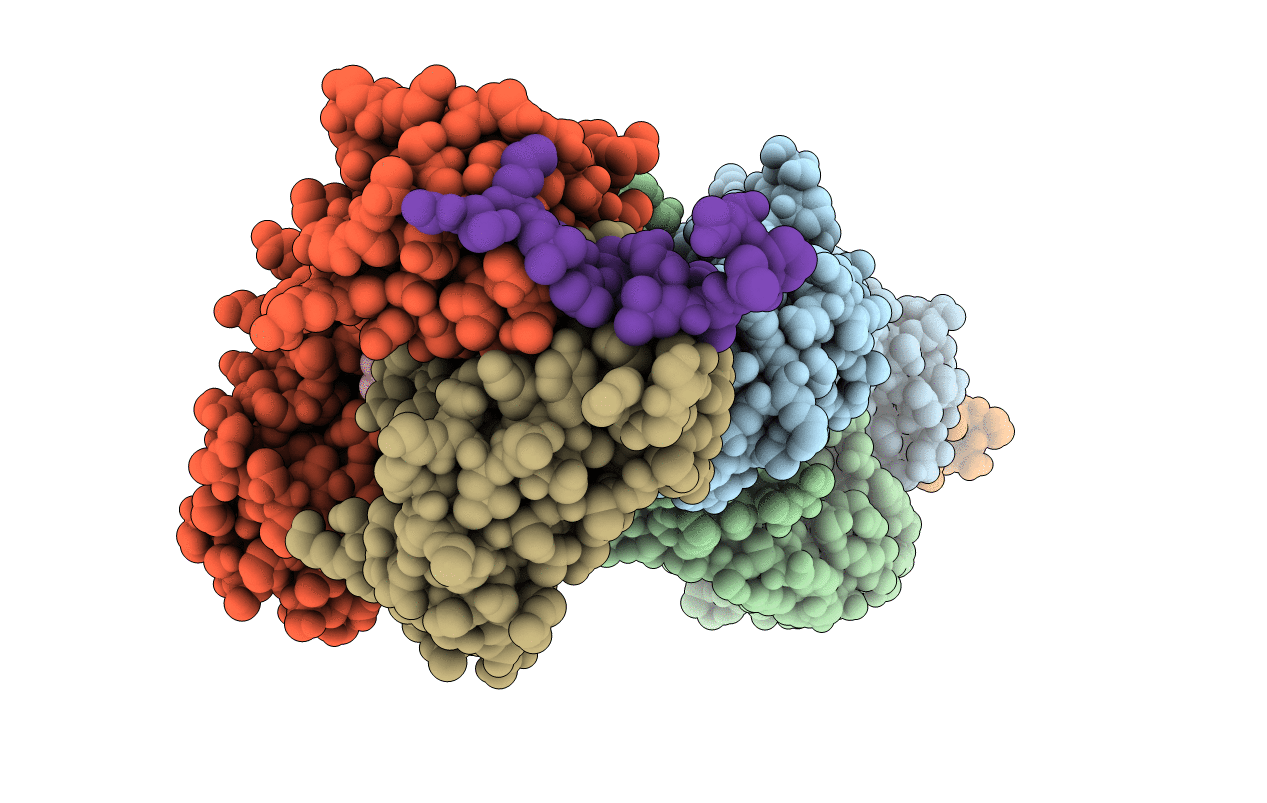 |
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-06-15 Classification: IMMUNE SYSTEM Ligands: TRS |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Resolution:15.00 Å Release Date: 2020-07-29 Classification: EXOCYTOSIS |
 |
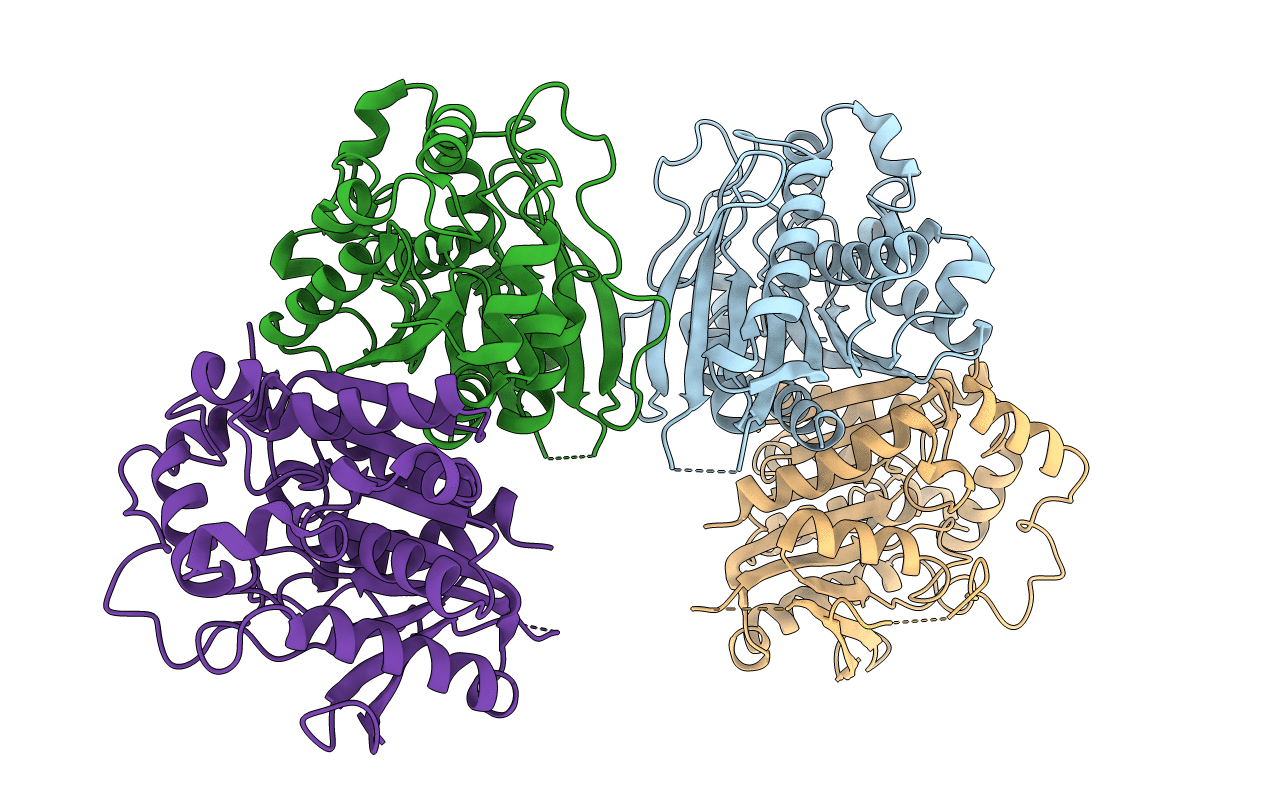
Crystal Structure Of Carboxyl Esterase 2 (Tmelest2) From Mycorrhizal Fungus Tuber Melanosporum
Organism: Tuber melanosporum mel28
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2017-08-23 Classification: HYDROLASE Ligands: TRT |
 |
Crystal Structure Of Carboxyl Esterase 2 (Tmelest2) From Mycorrhizal Fungus Tuber Melanosporum
Organism: Tuber melanosporum (strain mel28)
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2017-08-23 Classification: HYDROLASE |
 |
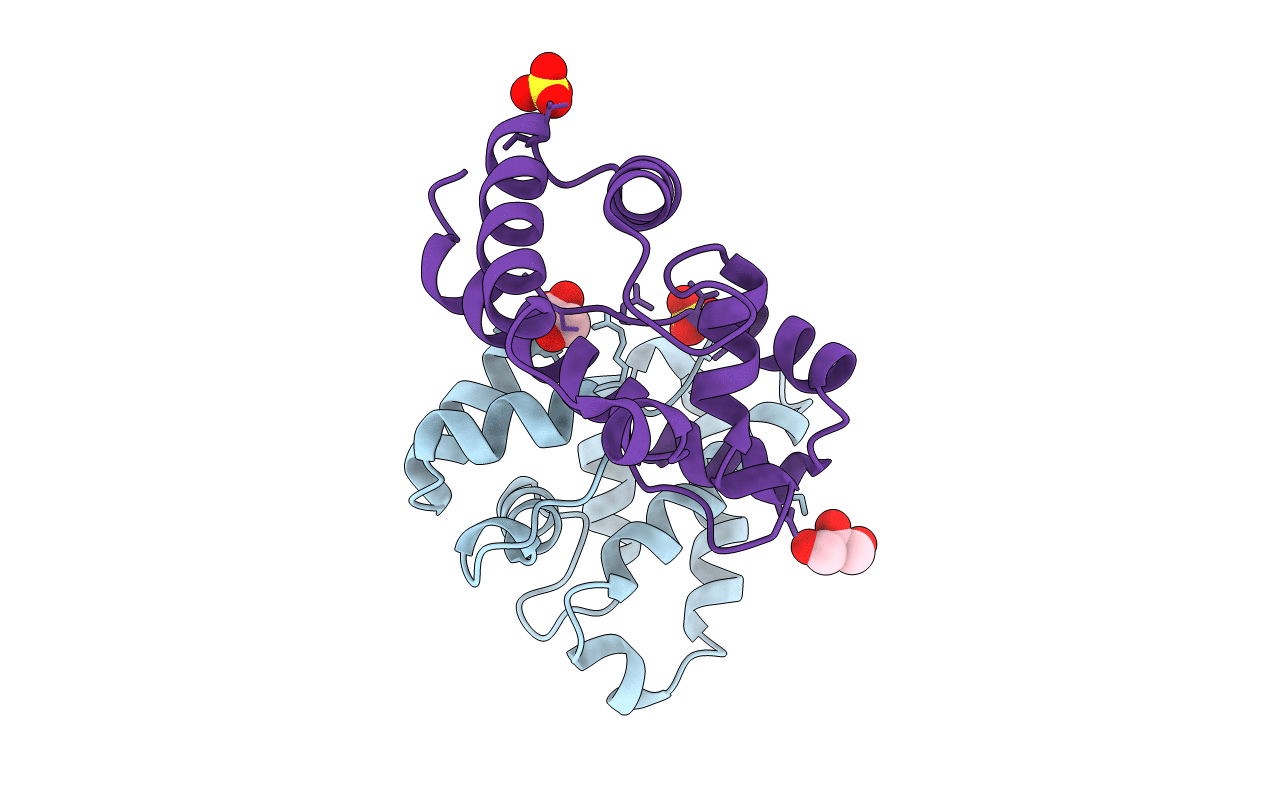
Organism: Helicobacter cinaedi
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2017-04-26 Classification: TRANSPORT PROTEIN Ligands: FE |
 |
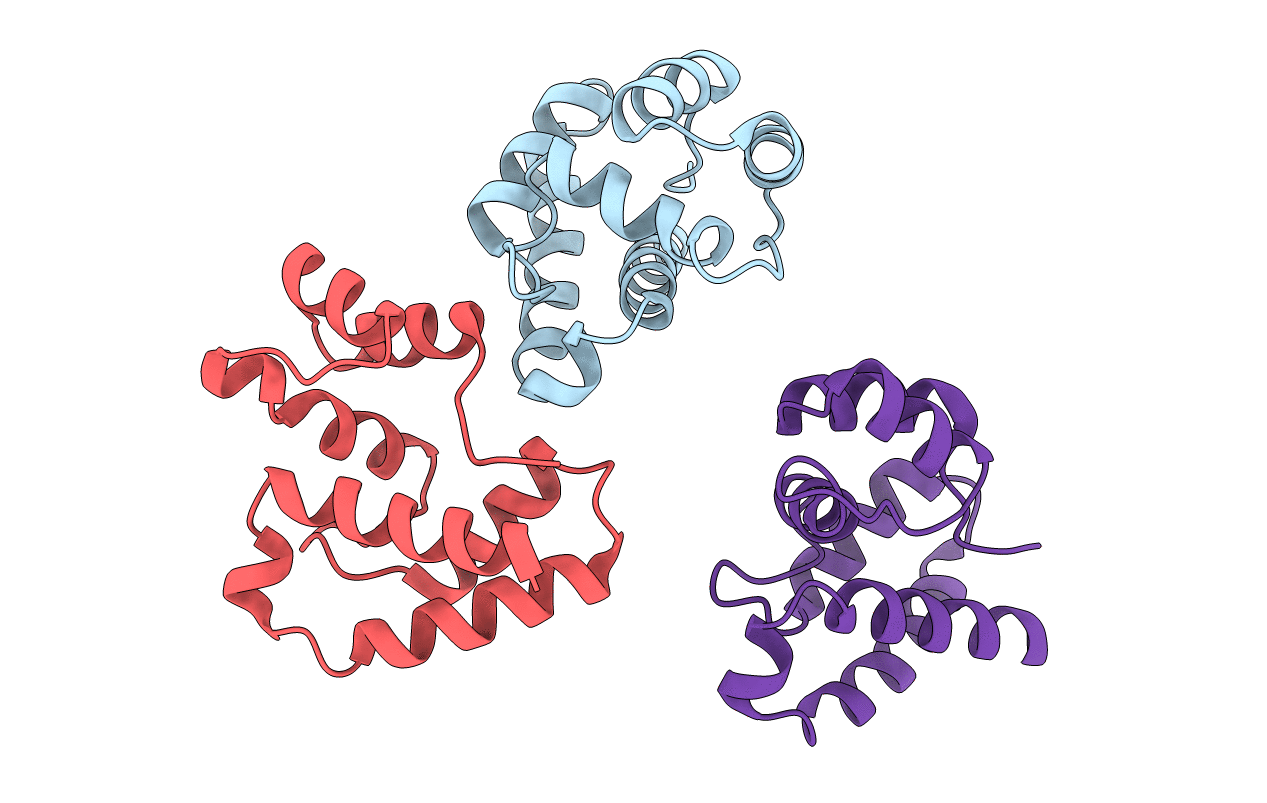
Organism: Megoura viciae
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2016-04-13 Classification: odorant binding protein Ligands: GOL, SO4 |
 |
Organism: Nasonovia ribisnigri
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2016-04-13 Classification: odorant binding protein |
 |
Organism: Nostoc
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-09-11 Classification: ELECTRON TRANSPORT Ligands: HEC |
 |
Organism: Nostoc
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-09-11 Classification: ELECTRON TRANSPORT Ligands: HEC |
 |
Organism: Nostoc
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-09-11 Classification: ELECTRON TRANSPORT Ligands: HEC |
 |
Organism: Tuber borchii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-12-12 Classification: HYDROLASE Ligands: ACT, SCN |
 |
Crystal Structure Of The Phosphorylation-Site Double Mutant S431E/T432E Of The Kaic Circadian Clock Protein
Organism: Synechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2011-09-21 Classification: TRANSFERASE Ligands: ATP, MG |
 |
Crystal Structures Of The G81A Mutant Of The Active Chimera Of (S)-Mandelate Dehydrogenase And Its Complex With Two Of Its Substrates
Organism: Pseudomonas putida, Spinacia oleracea
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2009-12-22 Classification: OXIDOREDUCTASE Ligands: FMN, MES |
 |
Organism: Paracoccus versutus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2008-12-30 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2008-03-18 Classification: ELECTRON TRANSPORT Ligands: CU, NI, 144, OPP |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2007-11-27 Classification: ELECTRON TRANSPORT Ligands: CU |
 |
Crystal Structure Of An Electron Transfer Complex Between Aromatic Amine Dehydrogenase And Azurin From Alcaligenes Faecalis (Form 3)
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2006-11-21 Classification: OXIDOREDUCTASE/electron transport Ligands: CU |
 |
Crystal Structure Of An Electron Transfer Complex Between Aromatic Amine Dephydrogenase And Azurin From Alcaligenes Faecalis (Form 1)
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2006-11-21 Classification: OXIDOREDUCTASE/electron transport Ligands: CU |
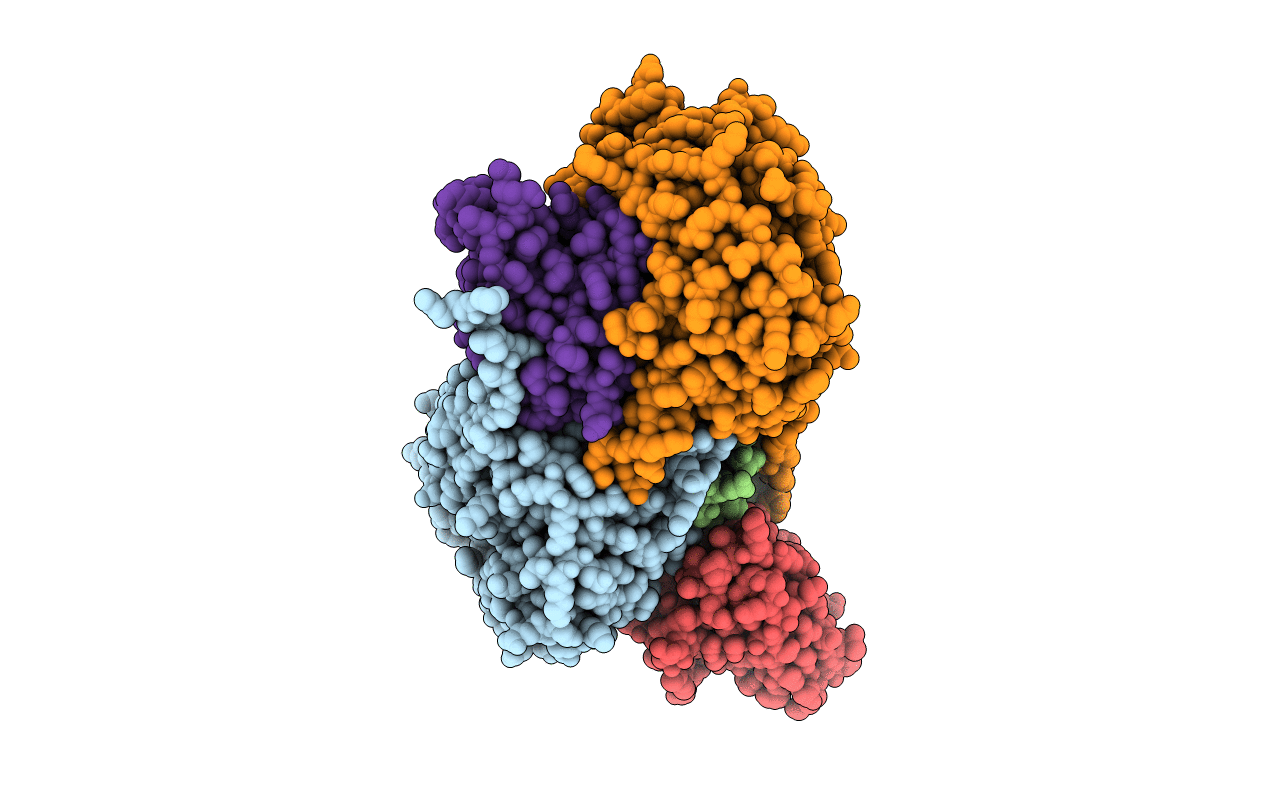 |
Crystal Structure Of An Electron Transfer Complex Between Aromatic Amine Dephydrogenase And Azurin From Alcaligenes Faecalis (Form 2)
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2006-11-21 Classification: OXIDOREDUCTASE/electron transport Ligands: CU |
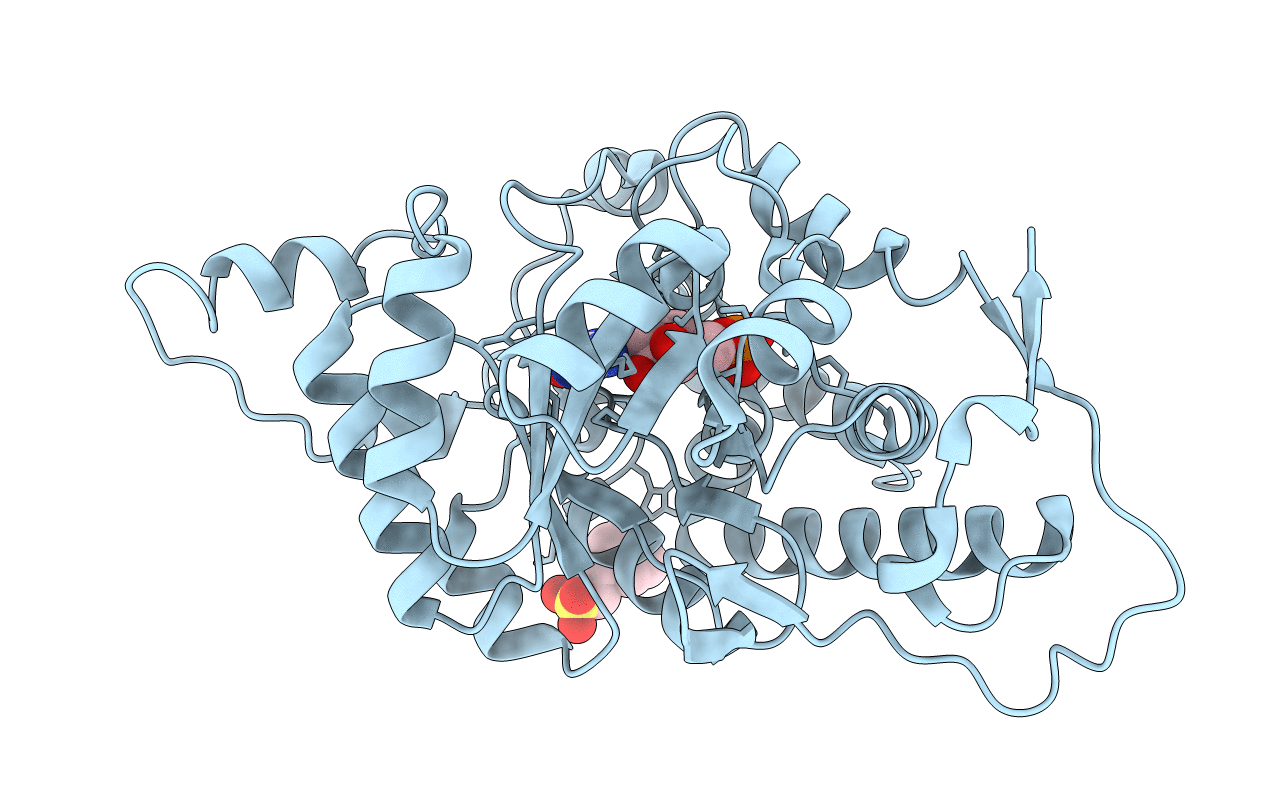 |
Crystal Structure Of The G81A Mutant Of The Active Chimera Of (S)-Mandelate Dehydrogenase
Organism: Pseudomonas putida, Spinacia oleracea
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-07-11 Classification: OXIDOREDUCTASE Ligands: FMN, MES |

