Search Count: 13
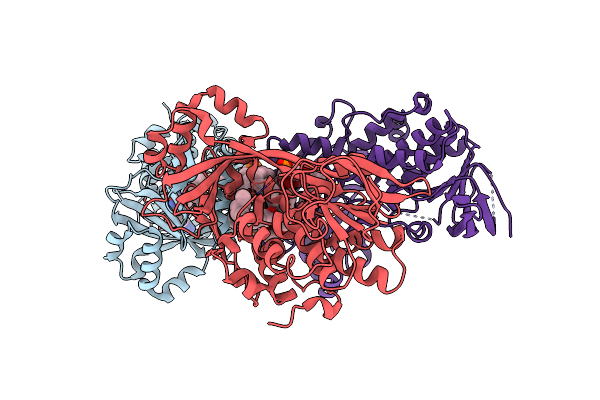 |
Organism: Neobacillus niacini
Method: X-RAY DIFFRACTION Resolution:3.14 Å Release Date: 2024-09-18 Classification: OXIDOREDUCTASE Ligands: FAD, DR9 |
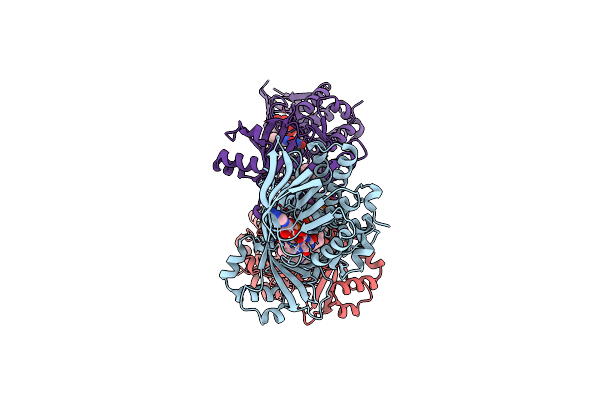 |
Organism: Neobacillus niacini
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: CYTOSOLIC PROTEIN Ligands: FAD, DR9 |
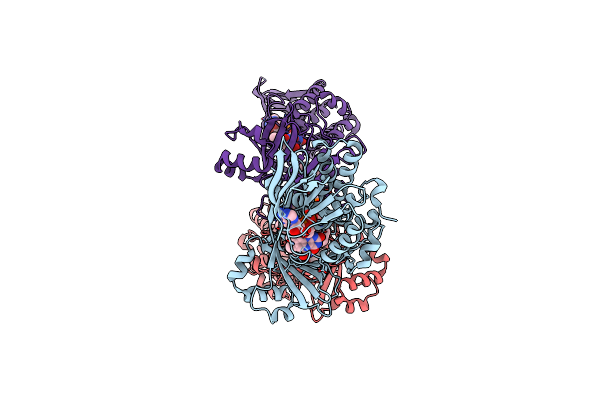 |
Organism: Neobacillus niacini
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: CYTOSOLIC PROTEIN Ligands: FAD, WTQ, DR9 |
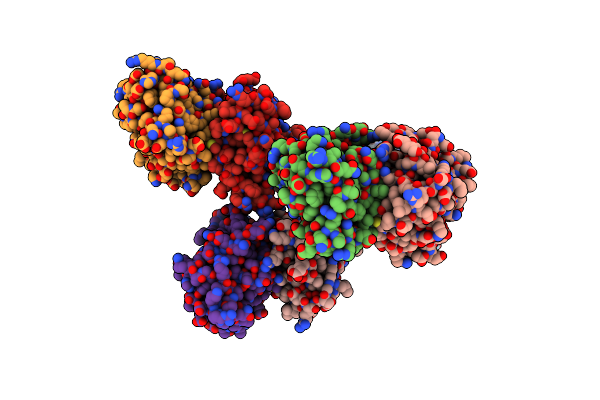 |
The Structure Of Ligand Binding Domain Of Lasr In Complex With Tp-1 Homolog, Compound 10
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-08-08 Classification: signaling protein/agonist Ligands: FXD |
 |
The Structure Of Ligand Binding Domain Of Lasr In Complex With Tp-1 Homolog, Compound 11
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-08-08 Classification: signaling protein/agonist Ligands: FXS |
 |
The Structure Of Ligand Binding Domain Of Lasr In Complex With Tp-1 Homolog, Compound 12
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2018-08-08 Classification: signaling protein/agonist Ligands: FYD, HIS |
 |
The Structure Of Ligand Binding Domain Of Lasr In Complex With Tp-1 Homolog, Compound 13
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-08-08 Classification: signaling protein/agonist Ligands: FY7 |
 |
The Structure Of Ligand Binding Domain Of Lasr In Complex With Tp-1 Homolog, Compound 14
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2018-08-08 Classification: signaling protein/agonist Ligands: FY4 |
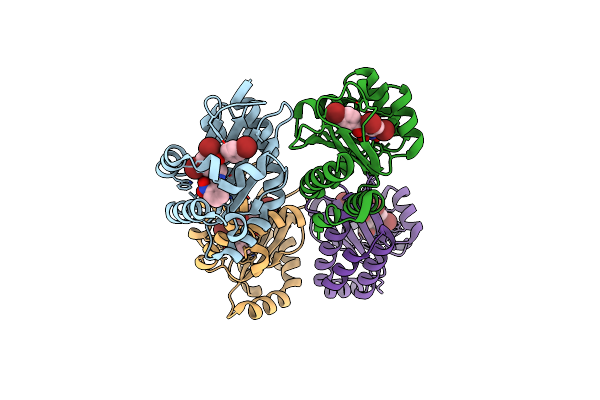 |
The Structure Of Ligand Binding Domain Of Lasr In Complex With Tp-1 Homolog, Compound 15
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-08-08 Classification: signaling protein/agonist Ligands: TX7 |
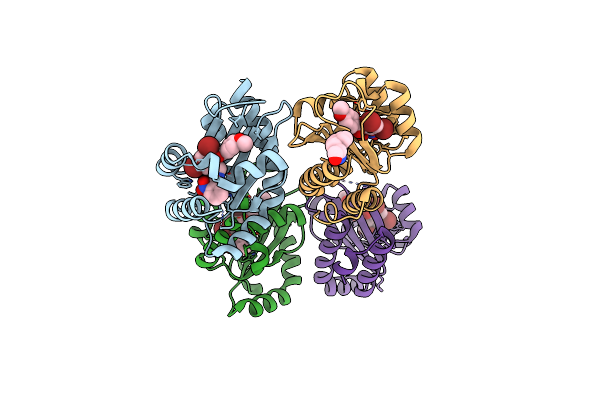 |
The Structure Of Ligand Binding Domain Of Lasr In Complex With Tp-1 Homolog, Compound 16
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2018-08-08 Classification: signaling protein/agonist Ligands: FXJ, PHE |
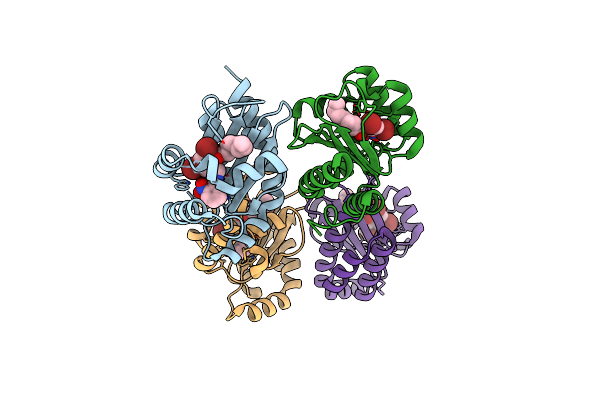 |
The Structure Of Ligand Binding Domain Of Lasr In Complex With Tp-1 Homolog, Compound 17
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2018-08-08 Classification: signaling protein/agonist Ligands: FX7 |
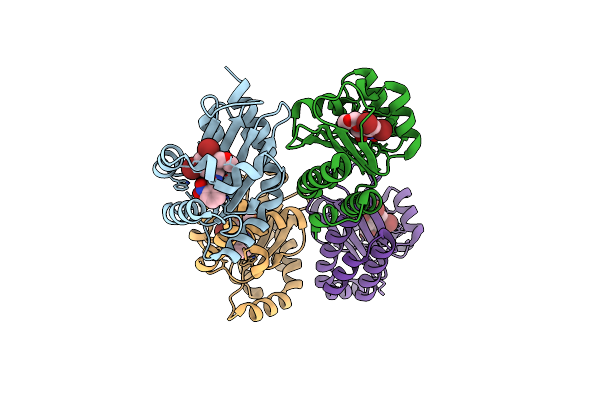 |
The Structure Of Ligand Binding Domain Of Lasr In Complex With Tp-1 Homolog, Compound 19
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2018-08-08 Classification: signaling protein/agonist Ligands: FY1 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-04-17 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: SAM, 1L8, UNX |

