Search Count: 11
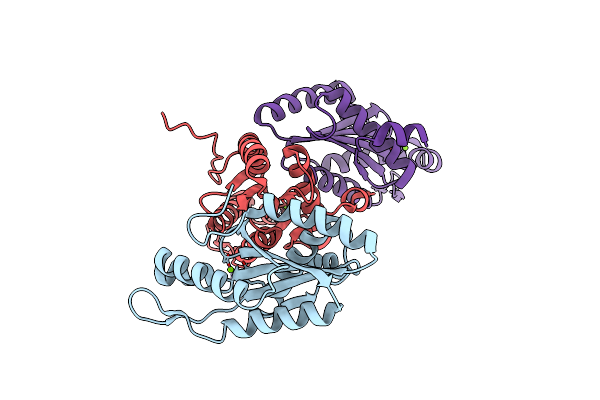 |
Structure Of A Delta-N Mutant - E232Start - Of Pa1120 (Tpbb Or Yfin) From Pseudomonas Aeruginosa (Pao1) Comprising Only The Ggdef Domain
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-07-07 Classification: LIGASE Ligands: MG |
 |
Organism: Salmonella typhimurium (strain sl1344)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-03-11 Classification: STRUCTURAL PROTEIN |
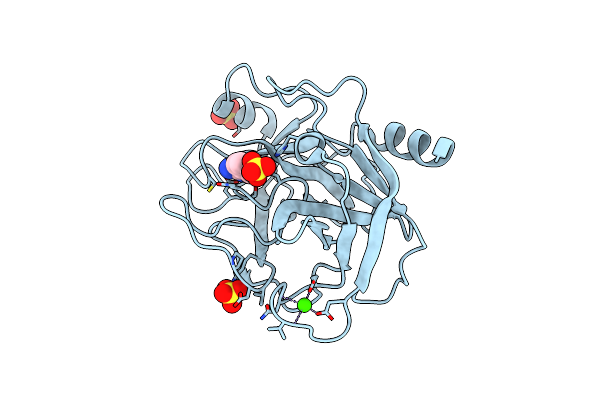 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-02-07 Classification: HYDROLASE Ligands: CA, BEN, SO4 |
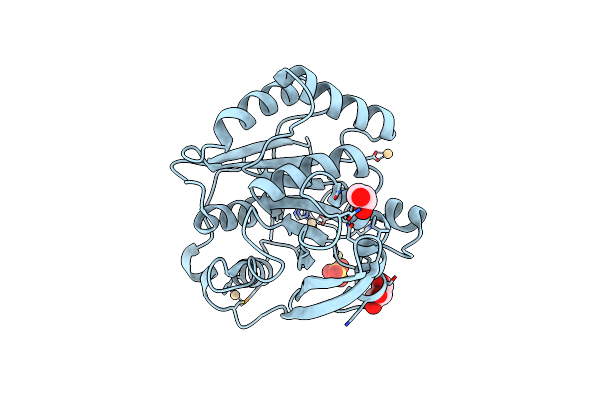 |
Structure Of Fae Solved By Sad From Data Collected At The Peak Of The Selenium Absorption Edge On Id30B
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-02-07 Classification: HYDROLASE Ligands: CD, GOL, SO4 |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2018-02-07 Classification: PLANT PROTEIN Ligands: TLA, GOL |
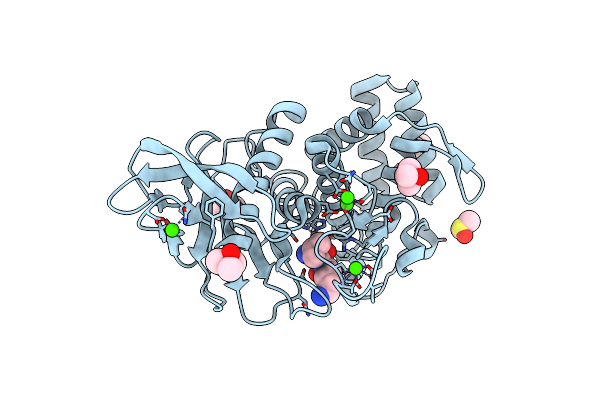 |
Structure Of Thermolysin Solved From Sad Data Collected At The Peak Of The Zn Absorption Edge On Id30B
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2018-01-31 Classification: HYDROLASE Ligands: ZN, CA, DMS, TMO, VAL, LYS |
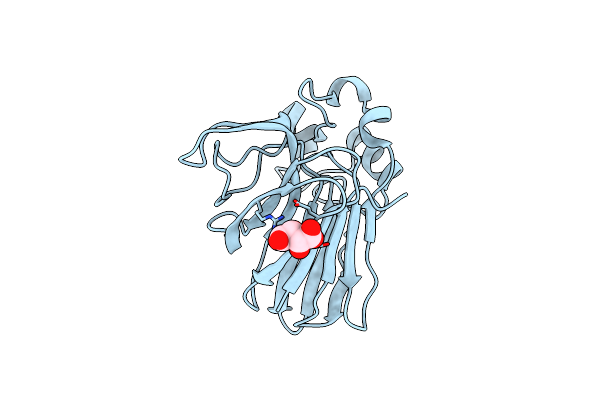 |
Structure Of Thaumatin Collected From An In Situ Crystal Collected On Id30B At 12.7 Kev.
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-01-31 Classification: PLANT PROTEIN Ligands: TLA |
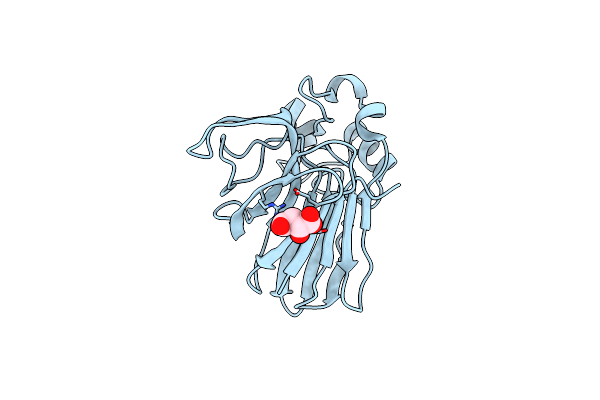 |
Structure Of Thaumatin Collected From An In Situ Crystal On Id30B At 17.5 Kev.
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-01-31 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2009-12-22 Classification: SIGNALING PROTEIN Ligands: GNP, MG, CA |
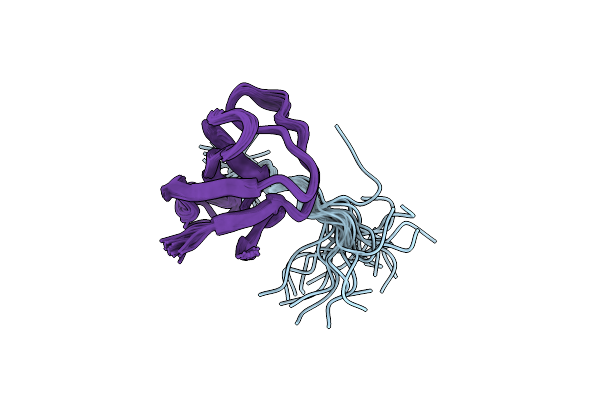 |
Nmr Study Of The Sh3 Domain From Fyn Proto-Oncogene Tyrosine Kinase Complexed With The Synthetic Peptide P2L Corresponding To Residues 91-104 Of The P85 Subunit Of Pi3-Kinase, Family Of 25 Structures
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 1998-02-25 Classification: COMPLEX (PHOSPHOTRANSFERASE/PEPTIDE) |
 |
Nmr Study Of The Sh3 Domain From Fyn Proto-Oncogene Tyrosine Kinase Kinase Complexed With The Synthetic Peptide P2L Corresponding To Residues 91-104 Of The P85 Subunit Of Pi3-Kinase, Minimized Average (Probmap) Structure
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 1998-02-25 Classification: COMPLEX (PHOSPHOTRANSFERASE/PEPTIDE) |

